
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,590,730 – 20,590,829 |
| Length | 99 |
| Max. P | 0.512088 |

| Location | 20,590,730 – 20,590,829 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.95 |
| Shannon entropy | 0.41467 |
| G+C content | 0.53402 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -18.47 |
| Energy contribution | -20.58 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.512088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
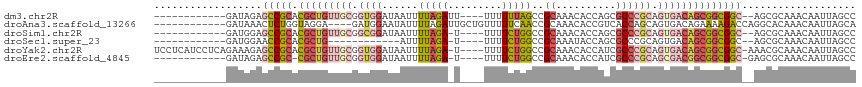
>dm3.chr2R 20590730 99 - 21146708 ------------GAUAGAGCCGCACGCUGUUGCGGUGGAUAAUUUUAGAUU----UUUUUUAGCCGCAAACACCAGCGCCCGCAGUGACAGCGGCGGC--AGCGCAAACAAUUAGCC ------------......(((((.((((((..(.((((......(((((..----...))))).(((........))).)))).)..)))))))))))--................. ( -35.50, z-score = -2.45, R) >droAna3.scaffold_13266 7182737 101 - 19884421 ------------GAUAAACUCUGGUAGGA----GAUGGAAUAUUUUAGAUUGCUGUUUUUCAACCGCAAACACCGUCACCAGCAGUGACAGAAAAGACCAGGCACAAACAAUUAGCA ------------.......((((((.((.----..((((....))))(.((((.(((....))).)))).).))(((((.....))))).......))))))............... ( -18.30, z-score = -0.06, R) >droSim1.chr2R 19046273 98 - 19596830 ------------GAUGGAGCCGCACGCUGUUGCGGCGGAUAAUUUUAGA-U----UUUUCUGGCCGCAAACACCAGCGCCCGCAGUGACAGCGGCGGC--AGCGCAAACAAUUAGCC ------------......(((((.((((((..(.((((......(((((-.----...))))).(((........))).)))).)..)))))))))))--................. ( -39.60, z-score = -2.20, R) >droSec1.super_23 457555 86 - 989336 ------------GAUGGAACCGCACGCUG------------AUUUUAGA-U----UUUUCUGGCCGCAAAUACCAGCGCCCGCAGUGACAGCGGCGGC--AGCGCAAACAAUUAGCC ------------..(((..((((.(((((------------.....(((-.----...)))((.(((........))).)).......))))))))).--..).))........... ( -23.30, z-score = 0.31, R) >droYak2.chr2R 20558191 111 - 21139217 UCCUCAUCCUCAGAAAGAGCCGCACGCUGUUGCGGUGGAUAAUUUUAGA-U----UUUUCUGGCCGCAAACACCAUCGCCCGCAGUGACAGCGGCGGC-AAACGCAAACAAUUAGCC ..................(((((.((((((..(.((((......(((((-.----...)))))..((..........)))))).)..)))))))))))-.................. ( -34.80, z-score = -1.65, R) >droEre2.scaffold_4845 21962412 98 - 22589142 ------------GAUAGAGCCGC-CGCUGUUGCGGUGGAUAAUUUUAGA-U----UUUUCUGGCCGCAAACACCAUCGCCCGCAGCGACGGCGGCGGC-GAGCGCAAACAAUUAGCC ------------......(((((-(((((((((.((((......(((((-.----...)))))..((..........)))))).))))))))))))))-.................. ( -40.10, z-score = -2.27, R) >consensus ____________GAUAGAGCCGCACGCUGUUGCGGUGGAUAAUUUUAGA_U____UUUUCUGGCCGCAAACACCAGCGCCCGCAGUGACAGCGGCGGC__AGCGCAAACAAUUAGCC ..................(((((.(((((((((.((((......(((((.........)))))..((..........)))))).))))))))))))))................... (-18.47 = -20.58 + 2.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:49:00 2011