| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,567,407 – 20,567,500 |
| Length | 93 |
| Max. P | 0.994181 |

| Location | 20,567,407 – 20,567,500 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.70 |
| Shannon entropy | 0.56231 |
| G+C content | 0.49891 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.74 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
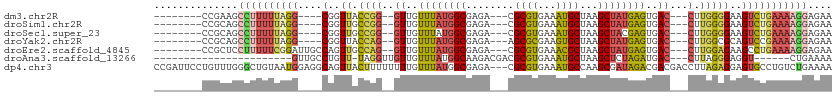
>dm3.chr2R 20567407 93 + 21146708 --------CCGAAGCCUUUUUAGG----CGGUUACCGG--GUUGUUUAUGGCGAGA---CGCGUGAAAUGCUAAGCUAUGAGUGAC---CUUGGGGAAGUCUGAAAAGGAGAA --------......((((((((((----(..(..((((--((..((((((((....---.((((...))))...))))))))..))---).)))..).))))))))))).... ( -36.80, z-score = -4.48, R) >droSim1.chr2R 19022801 93 + 19596830 --------CCGCAGCCUUUUUAGG----CGGUUGCCGG--GUUGUUUAUGGCGAGA---CGCGUGAAAUGCUAAGCUAUGAGUGAC---CUUGGGGAAGUCUGAAAAGGAGAA --------......((((((((((----(..((.((((--((..((((((((....---.((((...))))...))))))))..))---).))).)).))))))))))).... ( -38.20, z-score = -4.02, R) >droSec1.super_23 432932 93 + 989336 --------CCGCAGCCUUUUUAGG----CGGUUGCCGG--GUUGUUUAUGGCGAGA---CGCGUGAAAUGCUAAGCUACGAGUGAC---CUUGGGGAAGUCUGAAAAGGAGAA --------......((((((((((----(..((.((((--((..((..((((....---.((((...))))...))))..))..))---).))).)).))))))))))).... ( -35.10, z-score = -2.66, R) >droYak2.chr2R 20534265 93 + 21139217 --------CCGCAGCCUUUUUAGG----CGGUUACCAG--GUUGUUUAUGGCGAGA---AGCGCGAAGUGCUAAGCUAUGAGUGAC---CUUGGCGCAGUCCGAAAAGGAGAA --------......((((((..((----(.((..((((--((..((((((((....---(((((...)))))..))))))))..))---).))).)).)))..)))))).... ( -36.40, z-score = -2.87, R) >droEre2.scaffold_4845 21936824 97 + 22589142 --------CCGCUCCUUUUUCGGAUUGCCAGUUGCCAG--GUUGUUUAUGGCGAGA---CGCGUGAAACGCUAAGCUAUGAGUGAC---CUUGGAGAAGCCUGAAAAGGAGAA --------...(((((((((.((....(((......((--((..((((((((....---.(((.....)))...))))))))..))---))))).....)).))))))))).. ( -37.20, z-score = -3.69, R) >droAna3.scaffold_13266 7159064 80 + 19884421 -----------------------GUUGCCUGUU-UAGGUUGUUGUUUAUGGCAAGACGACGCGUGAAAUGCUAAGCUCUAGAUGAC---CUUAGGGAGGU------CUGAAAA -----------------------.......(((-(((((((((((((......)))))))((((...))))..))).))))))(((---(((...)))))------)...... ( -19.20, z-score = -0.37, R) >dp4.chr3 3919744 110 - 19779522 CCGAUUCCUGUUUGGGCUGUAAUGGAGGCAGUUACUUUUUUUUGUUUAUGGCGAGA---CGCGUGAAAUGCCAAGCGAUAGACGACGACCUUAGAGGAGUGCCUGUCUGAAAA ((((.......))))..........((((((.(((((((((((((((((.((....---.((((...))))...)).)))))))).......))))))))).))))))..... ( -27.71, z-score = -0.08, R) >consensus ________CCGCAGCCUUUUUAGG____CGGUUACCGG__GUUGUUUAUGGCGAGA___CGCGUGAAAUGCUAAGCUAUGAGUGAC___CUUGGGGAAGUCUGAAAAGGAGAA ..............((((((((((.......((.((((..((..((((((((........((((...))))...))))))))..))....)))).))..)))))))))).... (-14.86 = -15.74 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:56 2011