
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,565,550 – 20,565,650 |
| Length | 100 |
| Max. P | 0.573244 |

| Location | 20,565,550 – 20,565,650 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 64.30 |
| Shannon entropy | 0.60151 |
| G+C content | 0.51372 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -15.70 |
| Energy contribution | -13.24 |
| Covariance contribution | -2.46 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
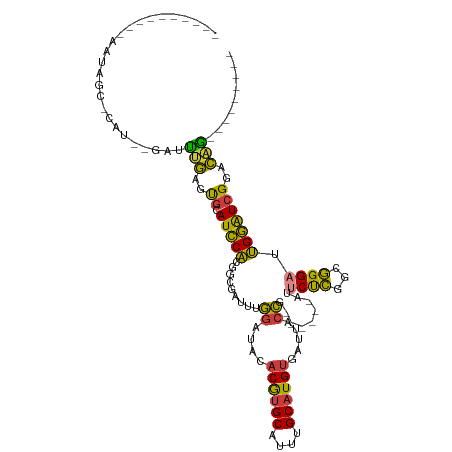
>dm3.chr2R 20565550 100 + 21146708 CUCGGUGGCCAAUGGC-CAU--GAUUUGAGUGCAUCCACGGCGAUUUGGAUACACAUGCAUUUGCAUGUGAUUACCG------AUUCUCGGCGGGAUUGGAUCGGACAG--------- ....((((((...)))-)))--((((..(.....(((.((.(((.((((...(((((((....)))))))....)))------)...))).))))))..))))......--------- ( -36.40, z-score = -2.07, R) >droPer1.super_4 900187 108 + 7162766 ----------AUUAGCUCGUUAGAUGCAUCCGCAUGCAUAUCGAUUGAGAUUUACGUGCAGUUGCAUGUGAUCACUGGGGGAGACACCGUUCUGGGAUGCAUCGUGUGCUCCAUGACC ----------...(((.((...(((((((((.((.((...........((((.((((((....))))))))))...((.(....).))))..))))))))))).)).)))........ ( -35.90, z-score = -0.85, R) >dp4.chr3 3917687 108 - 19779522 ----------AUUAGCUCGUUAGAUGCAUCCGCAUGCAUAUCGAUGGAGAUUUACGUGCAGUUGCAUGUGAUCACUGGGGGAGACACCGUUCUGGGAUGCAUCGUGUGCUCCAUGACC ----------...(((.((...(((((((((.((.((........((.((((.((((((....)))))))))).))((.(....).))))..))))))))))).)).)))........ ( -36.10, z-score = -0.64, R) >droEre2.scaffold_4845 21934995 96 + 22589142 ------CUCGCAGAGC-UAGCAGAUUUGAGUGCAUCCGCGGCGCUUUGGAUUAUCGCGCAUUUGCAUGUGAUCAGCG------AUUCUCGGCGGGAUCGGGUCGGACAG--------- ------(((((.(((.-..((.((((...(((((......((((...........))))...)))))..)))).)).------...))).)))))..............--------- ( -28.40, z-score = 1.36, R) >droYak2.chr2R 20532364 93 + 21139217 -------CUCAAUAGC-CGU--GAUCUGAGUGCAUCCACGGCGAUUUGGAUAAACGUGCACUUGCAUGUGAUUACCG------AUUCUCGACGGGAUUGGAUGGGACAG--------- -------((((...((-(((--(...((....))..))))))...........((((((....)))))).....(((------(((((....)))))))).))))....--------- ( -27.80, z-score = -1.01, R) >droSec1.super_23 431218 87 + 989336 -------------GGC-CAU--GAUUGGAGUGCAUCCACGGCGAUUUGGAUACACAUGCAUUUGCAUGUGAUUACCG------AUUCUCGGCGGGAUUGGAUCGGACAG--------- -------------..(-((.--...))).((.((((((((.(((.((((...(((((((....)))))))....)))------)...))).))....))))).).))..--------- ( -28.80, z-score = -1.59, R) >droSim1.chr2R 19021020 87 + 19596830 -------------GGC-CAU--GAUUUGAGUGCAUCCACGGCGAUUUGGAUACACAUGCAUUUGCAUGUUAUUACCG------AUUCUCAACGGGAUUGGAUCGGACAG--------- -------------.((-(.(--(...((....))..)).)))(((...((((.((((((....)))))))))).(((------(((((....)))))))))))......--------- ( -22.40, z-score = -0.77, R) >consensus __________AAUAGC_CAU__GAUUUGAGUGCAUCCACGGCGAUUUGGAUACACGUGCAUUUGCAUGUGAUUACCG______AUUCUCGGCGGGAUUGGAUCGGACAG_________ .........................(((..((.(((((.........((....((((((....)))))).....)).........((((...)))).)))))))..)))......... (-15.70 = -13.24 + -2.46)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:54 2011