| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,552,442 – 20,552,537 |
| Length | 95 |
| Max. P | 0.922718 |

| Location | 20,552,442 – 20,552,537 |
|---|---|
| Length | 95 |
| Sequences | 15 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Shannon entropy | 0.49630 |
| G+C content | 0.52804 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -18.79 |
| Energy contribution | -17.83 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
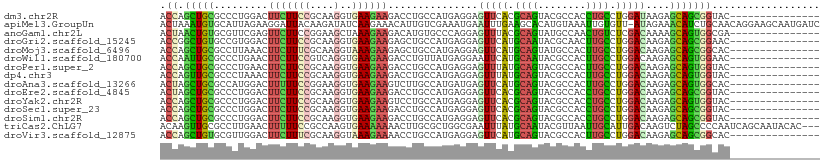
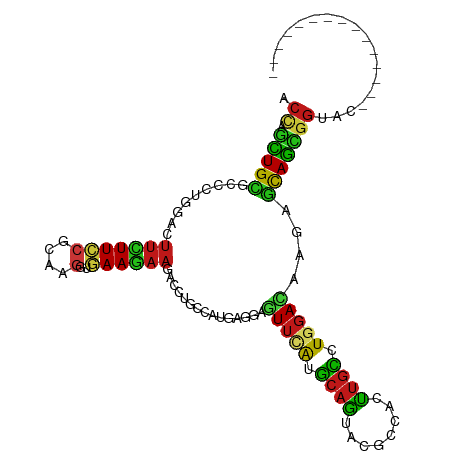
>dm3.chr2R 20552442 95 + 21146708 ACCAGCUGCGCCCUGGACUUCUUCCGCAAGGUGAAGAAGACCUGCCAUGAGGAGUUCACGCAGUACGCCACCUGCCUGGAUAAGAGCAGCGGUAC--------------- ....((((((...((((((((((..(((.(((.......))))))...))))))))))))))))..(((..((((((.....)).)))).)))..--------------- ( -34.90, z-score = -1.86, R) >apiMel3.GroupUn 18462822 109 + 399230636 ACUAAAUGUGCAUUAGAAGGAUUACAAGAUAUCAAGAAACAUUGUCGAAAUGAAUUUGAAGCACAUGUAAAUUGUGUU-AUAGAAACAUCUGCAACAGGAAGCAAUGAUC .....((((((.(((((..(((........)))......((((.....))))..))))).)))))).....((.((((-.((((....)))).)))).)).......... ( -18.40, z-score = -0.22, R) >anoGam1.chr2L 9367771 95 - 48795086 ACUAACUGUGCGUUCGAGUUCUUCCGGAAGCUAAAGAAGACAUGUGCCCAGGAGUUUACGCAGUAUGCCAACUGUCUCGACAAAAGCAGUGGCGA--------------- ........(((((.......((((.((..((............))..)).))))...)))))...(((((.((((..........))))))))).--------------- ( -21.20, z-score = 1.19, R) >droGri2.scaffold_15245 8387239 95 - 18325388 ACCGGCUGUGCCGUGGACUUCUUCCGCAAGGUGAAGAAGAGCUGCCAUGAGGAGUUCAUGCAAUACGCAACUUGCCUGGACAAGAGCAGCGGAAC--------------- .(((.((((..(((((.((((((((....))..)))))).....)))))....(((((.((((........)))).)))))....)))))))...--------------- ( -33.20, z-score = -1.30, R) >droMoj3.scaffold_6496 4088533 95 + 26866924 ACCAGCUGCGCCUUAAACUUCUUUCGCAAGGUAAAGAAGAGCUGCCAUGAGGAGUUCAUGCAGUAUGCCACUUGCCUGGACAAGAGCAGCGGCAC--------------- .((.(((((((......((((((((....))..)))))).(((((.(((((...))))))))))..))..((((......)))).)))))))...--------------- ( -30.40, z-score = -0.66, R) >droWil1.scaffold_180700 2189548 95 - 6630534 ACCAAUUGCGCCCUGAACUUCUUCCGUCAGGUGAAGAAGACCUGUUAUGAGGAAUUCAUGCAAUACGCCACUUGCCUGGACAAGAGCAGUGGAAC--------------- ....((((((...((((.((((((...(((((.......)))))....))))))))))))))))...(((((.((((.....)).)))))))...--------------- ( -28.90, z-score = -2.10, R) >droPer1.super_2 8065950 95 - 9036312 ACCAGCUGCGCCCUGAACUUCUUCCGCAAGGUGAAGAAGACCUGCCAUGAGGAGUUUAUGCAGUACGCCACUUGCCUGGACAAGAGCAGUGGUAC--------------- ....((((((...((((((((((..(((.(((.......))))))...))))))))))))))))..((((((.((((.....)).))))))))..--------------- ( -34.40, z-score = -2.40, R) >dp4.chr3 18978310 95 - 19779522 ACCAGUUGCGCCCUAAACUUCUUCCGCAAGGUGAAGAAGACCUGCCAUGAGGAGUUUAUGCAGUACGCCACUUGCCUGGACAAGAGCAGUGGUAC--------------- .....(((((...((((((((((..(((.(((.......))))))...)))))))))))))))...((((((.((((.....)).))))))))..--------------- ( -30.10, z-score = -1.51, R) >droAna3.scaffold_13266 7143384 95 + 19884421 ACUAGCUGCGCCAUGGACUUUUUCCGGAAGGUGAAGAAGUCUUGCCAUGAUGAGUUCAUGCAGUACGCCACUUGCCUGGACAAGAGCAGUGGCAC--------------- ....(((((((...(((((((((((....))..))))))))).)).((((.....)))))))))..((((((.((((.....)).))))))))..--------------- ( -37.40, z-score = -2.97, R) >droEre2.scaffold_4845 21921949 95 + 22589142 ACUAGCUGCGCCCUGGACUUCUUCCGCAAGGUGAAGAAGACCUGCCAUGAGGAGUUCACGCAGUACGCCACCUGCCUGGACAAGAGCAGCGGUAC--------------- ....((((((...((((((((((..(((.(((.......))))))...))))))))))))))))..(((..((((((.....)).)))).)))..--------------- ( -34.90, z-score = -1.67, R) >droYak2.chr2R 20519192 95 + 21139217 ACCAGCUGCGCCCUGGACUUCUUCCGCAAGGUGAAGAAGUCCUGCCAUGAGGAGUUCACGCAGUACGCCACCUGCCUGGACAAGAGCAGUGGUAC--------------- ((((.((((((...(((((((((((....))..))))))))).))........(((((.((((........)))).)))))....))))))))..--------------- ( -40.40, z-score = -3.04, R) >droSec1.super_23 418102 95 + 989336 ACCAGCUGCGCCCUGGACUUCUUCCGCAAGGUGAAGAAGACCUGCCAUGAGGAGUUCACGCAGUACGCCACCUGCCUGGACAAGAGCAGCGGUAC--------------- (((.(((((((...((.((((((((....))..)))))).)).))........(((((.((((........)))).)))))....))))))))..--------------- ( -36.50, z-score = -2.09, R) >droSim1.chr2R 19007643 95 + 19596830 ACCAGCUGCGCCCUGGACUUCUUCCGCAAGGUGAAGAAGACCUGCCAUGAGGAGUUCACGCAGUACGCCACCUGCCUGGACAAGAGCAGCGGUAC--------------- (((.(((((((...((.((((((((....))..)))))).)).))........(((((.((((........)))).)))))....))))))))..--------------- ( -36.50, z-score = -2.09, R) >triCas2.ChLG7 1882359 107 - 17478683 ACAAGUUGCGCCUUGAACUUUUUCCGCCAAGUGAAAAAAACUUGCGCUGGCGAAUUUAUGCAAUACGUUAAUUGCAUUGACAAGUCUAGCCCCAAUCAGCAAUACAC--- ....(((((...(((..........((.((((.......))))))((((((......(((((((......)))))))......).)))))..)))...)))))....--- ( -24.40, z-score = -1.36, R) >droVir3.scaffold_12875 1030240 95 + 20611582 ACCAGCUGUGCGUUGGACUUCUUUCGCAAGGUAAAGAAAACCUGCCAUGAGGAGUUCAUGCAGUACGCCACUUGCCUGGACAAGAGCAGCGGCAC--------------- ....(((((....((((((((((..(((.(((.......))))))...)))))))))).)))))..(((.((.((((.....)).)))).)))..--------------- ( -33.00, z-score = -1.42, R) >consensus ACCAGCUGCGCCCUGGACUUCUUCCGCAAGGUGAAGAAGACCUGCCAUGAGGAGUUCAUGCAGUACGCCACUUGCCUGGACAAGAGCAGCGGUAC_______________ .((.(((((.........(((((((....)..))))))...............(((((.((((........)))).)))))....))))))).................. (-18.79 = -17.83 + -0.96)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:52 2011