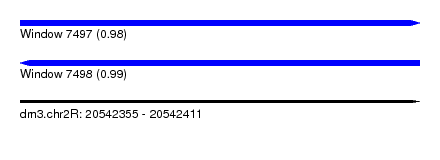
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,542,355 – 20,542,411 |
| Length | 56 |
| Max. P | 0.985784 |

| Location | 20,542,355 – 20,542,411 |
|---|---|
| Length | 56 |
| Sequences | 7 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 66.50 |
| Shannon entropy | 0.67729 |
| G+C content | 0.65531 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.47 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.983017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
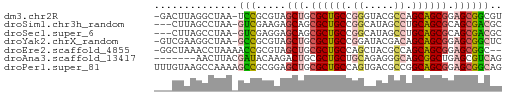
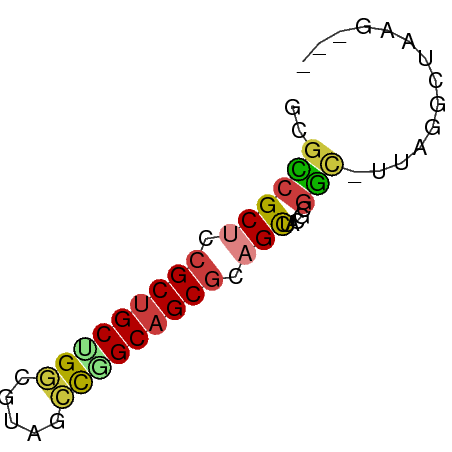
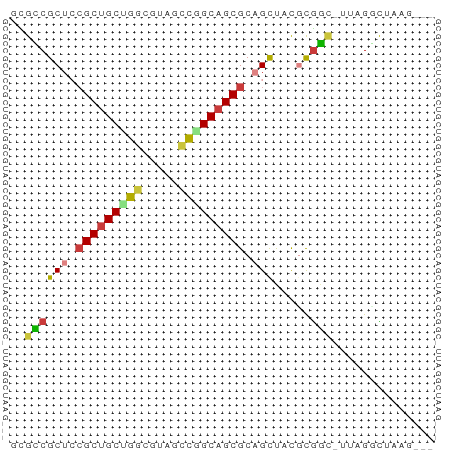
>dm3.chr2R 20542355 56 + 21146708 -GACUUAGGCUAA-UCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGCGU -......((....-.))((((.(((.((((((.((.....)).)))))).))).)))) ( -24.80, z-score = -0.54, R) >droSim1.chr3h_random 107942 54 - 1452968 ---CUUAGCCUAA-GUCGAAGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGC ---..........-((((.......(((((((.(((...))).)))))))..)))).. ( -22.50, z-score = -1.28, R) >droSec1.super_6 3669715 54 + 4358794 ---CUUAGCCUAA-GUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGC ---.....(((..-....))).((.(((((((.(((...))).))))))).))..... ( -23.50, z-score = -1.19, R) >droYak2.chrX_random 539170 56 + 1802292 -GUCGAAGGCUAA-GCCGCGUAGCUGCGCUGCCGGAUACGACAGCAGCGGAGCGGCUC -...........(-(((((....((((((((.((....)).)))).)))).)))))). ( -25.30, z-score = -1.23, R) >droEre2.scaffold_4855 7559 55 + 242605 -GGCUAAACCUAAAACCGCGUAGCUGCGCUGCCAGCUACGCCAGCAGCGGAGCGGC-- -..............(((((((((((......)))))))((.....))...)))).-- ( -18.60, z-score = 0.55, R) >droAna3.scaffold_13417 2534365 51 - 6960332 -------AACUUACGAUACAAGACUGCGCUGCUGCAGAGGGCAGCGGCUGAGCGUCAG -------..............(((.((((((((((.....))))))))...))))).. ( -19.00, z-score = -1.55, R) >droPer1.super_81 267500 58 - 277874 UUUGUAAGCCAAAAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGCGGCAG .......(((.....(....).(((.(((((((.(.....).))))))).)))))).. ( -25.90, z-score = -0.96, R) >consensus ___CUAAGCCUAA_GCCGCGGAGCUGCGCUGCCGGCUACGCCAGCAGCGGAGCGGCGC ..............(((.....(((.((((((.((.....)).)))))).)))))).. (-16.20 = -16.47 + 0.27)
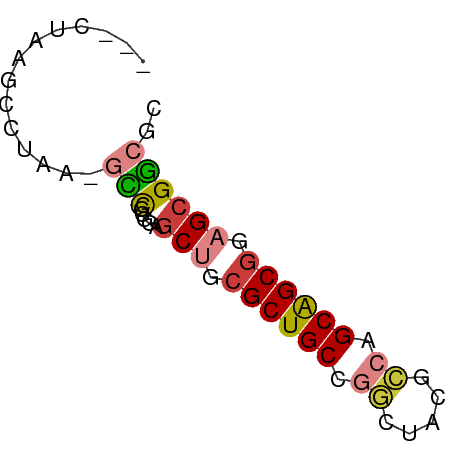
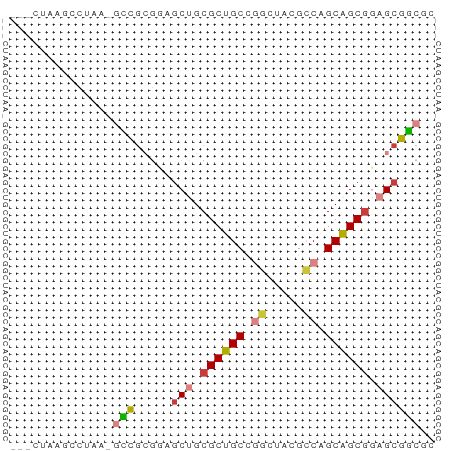
| Location | 20,542,355 – 20,542,411 |
|---|---|
| Length | 56 |
| Sequences | 7 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 66.50 |
| Shannon entropy | 0.67729 |
| G+C content | 0.65531 |
| Mean single sequence MFE | -22.63 |
| Consensus MFE | -18.08 |
| Energy contribution | -18.00 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20542355 56 - 21146708 ACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGA-UUAGCCUAAGUC- .(((.(((.(((((((((.....))))))))).)))...)))..-............- ( -24.60, z-score = -1.39, R) >droSim1.chr3h_random 107942 54 + 1452968 GCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCUUCGAC-UUAGGCUAAG--- ..(((((.(((((((.(((...))).))))))).)).....)))-..........--- ( -22.50, z-score = -0.50, R) >droSec1.super_6 3669715 54 - 4358794 GCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGAC-UUAGGCUAAG--- ..(((((.(((((((.(((...))).))))))).)).....)))-..........--- ( -22.50, z-score = -0.35, R) >droYak2.chrX_random 539170 56 - 1802292 GAGCCGCUCCGCUGCUGUCGUAUCCGGCAGCGCAGCUACGCGGC-UUAGCCUUCGAC- (((((((...((((((((((....)))))..)))))...)))))-))..........- ( -25.70, z-score = -2.09, R) >droEre2.scaffold_4855 7559 55 - 242605 --GCCGCUCCGCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGUUUAGCC- --((.(((.((((((..((...))..)))))).)))...))((((........))))- ( -24.00, z-score = -0.55, R) >droAna3.scaffold_13417 2534365 51 + 6960332 CUGACGCUCAGCCGCUGCCCUCUGCAGCAGCGCAGUCUUGUAUCGUAAGUU------- (((.((((.....(((((.....))))))))))))................------- ( -14.00, z-score = -0.15, R) >droPer1.super_81 267500 58 + 277874 CUGCCGCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUUUUGGCUUACAAA ((((.(((.(((((((((.....))))))))).)))...))))..((((.....)))) ( -25.10, z-score = -1.12, R) >consensus GCGCCGCUCCGCUGCUGGCGUAGCCGGCAGCGCAGCUACGCGGC_UUAGGCUAAG___ ..((((((.(((((((((.....))))))))).))).....))).............. (-18.08 = -18.00 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:50 2011