
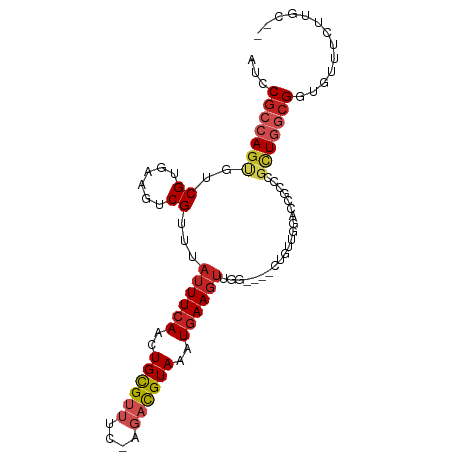
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,524,815 – 20,524,906 |
| Length | 91 |
| Max. P | 0.731346 |

| Location | 20,524,815 – 20,524,906 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.31 |
| Shannon entropy | 0.38484 |
| G+C content | 0.49363 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -15.35 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20524815 91 + 21146708 AUCCGCCAGUGUCGUGAAGUCGUUUAUUUCAACUGCGUUUC-AGACGUAAAUGAAGUUGG----CUUUUGGACCGCCCGCUGGCGGUGUUUCUUGC-- ..(((((((((((..(((((((...((((((..((((((..-.))))))..)))))))))----))))..))).....))))))))..........-- ( -31.50, z-score = -2.59, R) >droAna3.scaffold_13266 7119907 90 + 19884421 AUCCGCCAGUGUCGUGAAGUCGUUUAUUUCAACUGCGUUUU-AGACGUAAAUGAAGUUGG-----CGUUGGACCGCCCGCUGGCGGUGUCUGUUAC-- ..(((((((((..(((....(((..((((((..((((((..-.))))))..))))))..)-----))......))).)))))))))..........-- ( -32.40, z-score = -2.79, R) >droEre2.scaffold_4845 21901812 91 + 22589142 AUCCGCCAGUGUCGUGAAGUCGUUUAUUUCAACUGCGUUUC-AGACGUAAAUGAAGUUGG----CUUGUGGACCGCCCGCUGGCGGUGUUCCUUGC-- ..(((((((((..(((..(((((..((((((..((((((..-.))))))..))))))..)----).....)))))).)))))))))..........-- ( -30.60, z-score = -2.04, R) >droYak2.chr2R 20497684 91 + 21139217 AUCCGCCAGUGUCGUGAAGUCGUUUAUUUCAACUGCGUUUC-AGACGUAAAUGAAGUUGG----CUUUUGGACCGCCCGCUGGCGUUGUUUCUCGC-- ...((((((((((..(((((((...((((((..((((((..-.))))))..)))))))))----))))..))).....)))))))...........-- ( -28.40, z-score = -1.81, R) >droSec1.super_23 398023 91 + 989336 AUCCGCCAGUGUCGUGAAGUCGUUUAUUUCAACUGCGUUUC-AGACGUAAAUGAAGUUGG----CUGUUGGACCGCCCGCUGGCGGGGUUUCUUGC-- .((((((((((..(((..(((((..((((((..((((((..-.))))))..))))))..)----).....)))))).)))))))))).........-- ( -32.00, z-score = -1.91, R) >droSim1.chr2R 18987598 91 + 19596830 AUCCGCCAGUGUCGUGAAGUCGUUUAUUUCAACUGCGUUUC-AGACGUAAAUGAAGUUGG----CUGUUGGACCGCCCGCUGGCGGGGUUUCUUGC-- .((((((((((..(((..(((((..((((((..((((((..-.))))))..))))))..)----).....)))))).)))))))))).........-- ( -32.00, z-score = -1.91, R) >droGri2.scaffold_15245 8353653 89 - 18325388 AUCCGCCAGUGUCGUAAAGUCGUUUAUUUCAACUGUG--GCGAGACGUAAAUGAAGUUAA-----UGUCG--UCGUCGUUUUACGGUUGUCCUUGUUC ..(((.(((((..(((((....)))))..).)))).(--(((((((((.(((...))).)-----)))).--)))))......)))............ ( -16.90, z-score = 0.01, R) >droVir3.scaffold_12875 999366 87 + 20611582 AUCCGCCAGUGUCGUAAAGUCGUUUAUUUCAACUGUG--GCGAGACGUAAAUGAAGUUAA-----UGUCC--UCGUCGUUUUACGACUGUCCUUGC-- ....((.((.(((((((((.((((......)))...(--(((((((((.(((...))).)-----))).)--)))))))))))))))....)).))-- ( -21.00, z-score = -1.50, R) >droWil1.scaffold_180700 2133393 84 - 6630534 AUCCGCCAGCGUCGUGAAGUCGUUUAUUUCAACUGCGUUUUUAGAUGUAAGUAAA--UGA-----AGUUAAAU-----GGUGGAGACGGCUCUUCC-- .(((((((......(((..((((((((((((.(((......))).)).)))))))--)))-----..)))..)-----))))))............-- ( -22.80, z-score = -2.04, R) >droPer1.super_31 613926 96 + 935084 AUCCGCCAGCGUCGUGAAGUCGUUUAUUUCAACUGCGUUUUCAGACGUAAAUGAAGUUAAGUUACCGUUGGACCGCCCGCUGGCGGUGUCUCUUUC-- ..(((((((((..(((..(((......((((..((((((....))))))..))))((......)).....)))))).)))))))))..........-- ( -30.30, z-score = -2.38, R) >dp4.chr3 2603804 96 - 19779522 AUCCGCCAGCGUCGUGAAGUCGUUUAUUUCAACUGCGUUUUCAGACGUAAAUGAAGUUAAGUUACCGUUGGACCGCCCGCUGGCGGUGUCUCUUGC-- ..(((((((((..(((..(((......((((..((((((....))))))..))))((......)).....)))))).)))))))))..........-- ( -30.30, z-score = -1.92, R) >consensus AUCCGCCAGUGUCGUGAAGUCGUUUAUUUCAACUGCGUUUC_AGACGUAAAUGAAGUUGG____CUGUUGGACCGCCCGCUGGCGGUGUUUCUUGC__ ...(((((((..((......))...((((((..((((((....))))))..)))))).....................)))))))............. (-15.35 = -16.20 + 0.85)


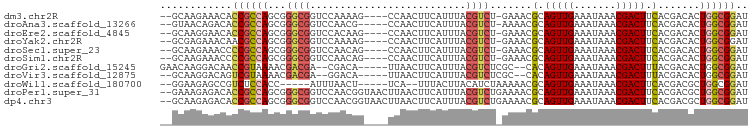
| Location | 20,524,815 – 20,524,906 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.31 |
| Shannon entropy | 0.38484 |
| G+C content | 0.49363 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -13.29 |
| Energy contribution | -13.31 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2R 20524815 91 - 21146708 --GCAAGAAACACCGCCAGCGGGCGGUCCAAAAG----CCAACUUCAUUUACGUCU-GAAACGCAGUUGAAAUAAACGACUUCACGACACUGGCGGAU --..........(((((((..(((.........)----))............((((-((.....(((((.......)))))))).))).))))))).. ( -25.30, z-score = -1.88, R) >droAna3.scaffold_13266 7119907 90 - 19884421 --GUAACAGACACCGCCAGCGGGCGGUCCAACG-----CCAACUUCAUUUACGUCU-AAAACGCAGUUGAAAUAAACGACUUCACGACACUGGCGGAU --..........(((((((.(((((......))-----))................-.....(.(((((.......))))).)....).))))))).. ( -26.70, z-score = -2.74, R) >droEre2.scaffold_4845 21901812 91 - 22589142 --GCAAGGAACACCGCCAGCGGGCGGUCCACAAG----CCAACUUCAUUUACGUCU-GAAACGCAGUUGAAAUAAACGACUUCACGACACUGGCGGAU --..........(((((((..(((.........)----))............((((-((.....(((((.......)))))))).))).))))))).. ( -25.30, z-score = -1.54, R) >droYak2.chr2R 20497684 91 - 21139217 --GCGAGAAACAACGCCAGCGGGCGGUCCAAAAG----CCAACUUCAUUUACGUCU-GAAACGCAGUUGAAAUAAACGACUUCACGACACUGGCGGAU --...........((((((..(((.........)----))............((((-((.....(((((.......)))))))).))).))))))... ( -22.20, z-score = -0.63, R) >droSec1.super_23 398023 91 - 989336 --GCAAGAAACCCCGCCAGCGGGCGGUCCAACAG----CCAACUUCAUUUACGUCU-GAAACGCAGUUGAAAUAAACGACUUCACGACACUGGCGGAU --..........(((((((..(((.(.....).)----))............((((-((.....(((((.......)))))))).))).))))))).. ( -25.20, z-score = -1.55, R) >droSim1.chr2R 18987598 91 - 19596830 --GCAAGAAACCCCGCCAGCGGGCGGUCCAACAG----CCAACUUCAUUUACGUCU-GAAACGCAGUUGAAAUAAACGACUUCACGACACUGGCGGAU --..........(((((((..(((.(.....).)----))............((((-((.....(((((.......)))))))).))).))))))).. ( -25.20, z-score = -1.55, R) >droGri2.scaffold_15245 8353653 89 + 18325388 GAACAAGGACAACCGUAAAACGACGA--CGACA-----UUAACUUCAUUUACGUCUCGC--CACAGUUGAAAUAAACGACUUUACGACACUGGCGGAU ............((.......((.((--((...-----.............))))))((--((..(((((((........))).))))..)))))).. ( -14.59, z-score = -0.60, R) >droVir3.scaffold_12875 999366 87 - 20611582 --GCAAGGACAGUCGUAAAACGACGA--GGACA-----UUAACUUCAUUUACGUCUCGC--CACAGUUGAAAUAAACGACUUUACGACACUGGCGGAU --....((((.((((.....))))((--((...-----....))))......))))(((--((..(((((((........))).))))..)))))... ( -21.80, z-score = -2.32, R) >droWil1.scaffold_180700 2133393 84 + 6630534 --GGAAGAGCCGUCUCCACC-----AUUUAACU-----UCA--UUUACUUACAUCUAAAAACGCAGUUGAAAUAAACGACUUCACGACGCUGGCGGAU --.......(((((..(...-----.(((((((-----.(.--(((............))).).))))))).....((......))..)..))))).. ( -10.20, z-score = 0.83, R) >droPer1.super_31 613926 96 - 935084 --GAAAGAGACACCGCCAGCGGGCGGUCCAACGGUAACUUAACUUCAUUUACGUCUGAAAACGCAGUUGAAAUAAACGACUUCACGACGCUGGCGGAU --..........(((((((((.(((.((..(((..................)))..))...)))(((((.......)))))......))))))))).. ( -30.27, z-score = -2.74, R) >dp4.chr3 2603804 96 + 19779522 --GCAAGAGACACCGCCAGCGGGCGGUCCAACGGUAACUUAACUUCAUUUACGUCUGAAAACGCAGUUGAAAUAAACGACUUCACGACGCUGGCGGAU --..........(((((((((.(((.((..(((..................)))..))...)))(((((.......)))))......))))))))).. ( -30.27, z-score = -2.52, R) >consensus __GCAAGAAACACCGCCAGCGGGCGGUCCAACAG____CCAACUUCAUUUACGUCU_GAAACGCAGUUGAAAUAAACGACUUCACGACACUGGCGGAU ............((((((...((((..........................)))).......(.(((((.......))))).).......)))))).. (-13.29 = -13.31 + 0.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:49 2011