
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,877,778 – 3,877,831 |
| Length | 53 |
| Max. P | 0.964726 |

| Location | 3,877,778 – 3,877,831 |
|---|---|
| Length | 53 |
| Sequences | 6 |
| Columns | 56 |
| Reading direction | forward |
| Mean pairwise identity | 81.73 |
| Shannon entropy | 0.34141 |
| G+C content | 0.49579 |
| Mean single sequence MFE | -12.32 |
| Consensus MFE | -10.13 |
| Energy contribution | -9.72 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
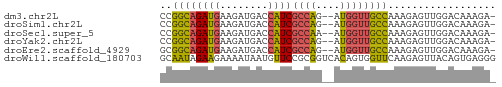
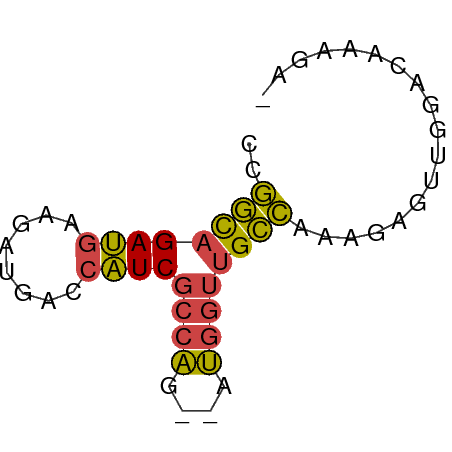
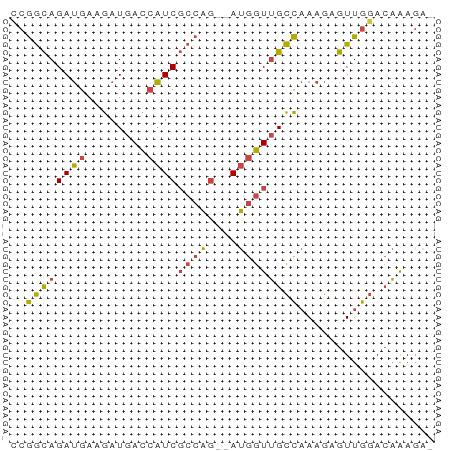
>dm3.chr2L 3877778 53 + 23011544 CCGGCAGAUGAAGAUGACCAUCGCCAG--AUGGUUGCCAAAGAGUUGGACAAAGA- (((((.......(..(((((((....)--))))))..).....))))).......- ( -13.80, z-score = -2.02, R) >droSim1.chr2L 3833063 53 + 22036055 CCGGCAGAUGAAGAUGACCAUCGCCAG--AUGGUUGCCAAAGAGUUGGACAAAGA- (((((.......(..(((((((....)--))))))..).....))))).......- ( -13.80, z-score = -2.02, R) >droSec1.super_5 1979819 53 + 5866729 CCGGCAGAUGAAGAUGACCAUCGCCAA--AUGGUUGCCAAAGAGUUGGACAAAGA- ..(((.((((........)))))))..--....(((((((....)))).)))...- ( -12.90, z-score = -1.65, R) >droYak2.chr2L 3886117 53 + 22324452 CCGGCAGAUGAAGAUGACCAUCGCCAG--AUGGUUGCCAAAGAGUUGGACAAAGA- (((((.......(..(((((((....)--))))))..).....))))).......- ( -13.80, z-score = -2.02, R) >droEre2.scaffold_4929 3929716 53 + 26641161 GCGGCAGAUGAAGAUGACCAUCGCCAG--AUGGUUGCCAAAGAGUUGGACAAAGA- ..(((.((((........)))))))..--....(((((((....)))).)))...- ( -12.90, z-score = -1.42, R) >droWil1.scaffold_180703 1789756 56 + 3946847 GCAAUAGAAGAAAAUAAUGUUCCGCGGUCACAGUGGUUCAAGAGUUACAGUGAGGG .................((..((((.......))))..))................ ( -6.70, z-score = 0.55, R) >consensus CCGGCAGAUGAAGAUGACCAUCGCCAG__AUGGUUGCCAAAGAGUUGGACAAAGA_ ..((((((((........))))((((....)))))))).................. (-10.13 = -9.72 + -0.41)
| Location | 3,877,778 – 3,877,831 |
|---|---|
| Length | 53 |
| Sequences | 6 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 81.73 |
| Shannon entropy | 0.34141 |
| G+C content | 0.49579 |
| Mean single sequence MFE | -12.77 |
| Consensus MFE | -9.33 |
| Energy contribution | -8.62 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
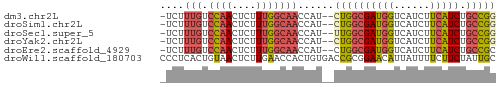
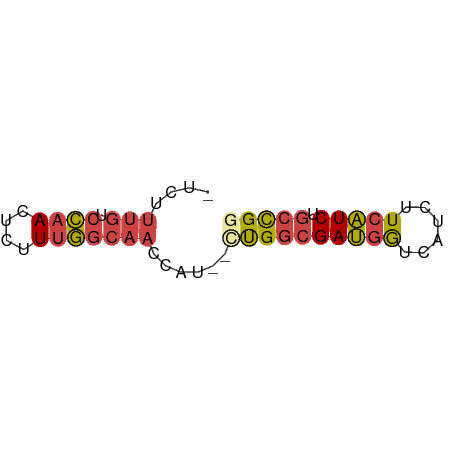
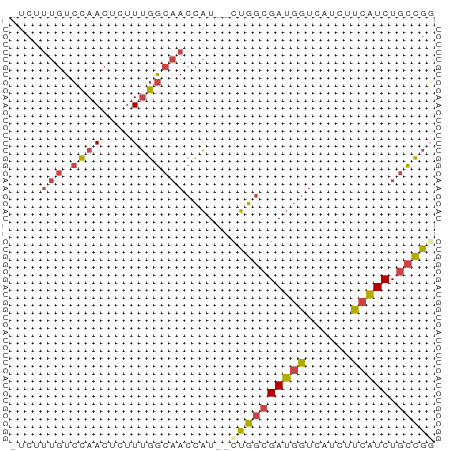
>dm3.chr2L 3877778 53 - 23011544 -UCUUUGUCCAACUCUUUGGCAACCAU--CUGGCGAUGGUCAUCUUCAUCUGCCGG -...(((.((((....)))))))....--((((((((((......))))).))))) ( -15.40, z-score = -3.07, R) >droSim1.chr2L 3833063 53 - 22036055 -UCUUUGUCCAACUCUUUGGCAACCAU--CUGGCGAUGGUCAUCUUCAUCUGCCGG -...(((.((((....)))))))....--((((((((((......))))).))))) ( -15.40, z-score = -3.07, R) >droSec1.super_5 1979819 53 - 5866729 -UCUUUGUCCAACUCUUUGGCAACCAU--UUGGCGAUGGUCAUCUUCAUCUGCCGG -...(((.((((....)))))))....--.(((((((((......))))).)))). ( -13.70, z-score = -2.27, R) >droYak2.chr2L 3886117 53 - 22324452 -UCUUUGUCCAACUCUUUGGCAACCAU--CUGGCGAUGGUCAUCUUCAUCUGCCGG -...(((.((((....)))))))....--((((((((((......))))).))))) ( -15.40, z-score = -3.07, R) >droEre2.scaffold_4929 3929716 53 - 26641161 -UCUUUGUCCAACUCUUUGGCAACCAU--CUGGCGAUGGUCAUCUUCAUCUGCCGC -...(((.((((....)))))))....--..((((((((......))))).))).. ( -13.50, z-score = -2.28, R) >droWil1.scaffold_180703 1789756 56 - 3946847 CCCUCACUGUAACUCUUGAACCACUGUGACCGCGGAACAUUAUUUUCUUCUAUUGC .......................((((....))))..................... ( -3.20, z-score = 0.84, R) >consensus _UCUUUGUCCAACUCUUUGGCAACCAU__CUGGCGAUGGUCAUCUUCAUCUGCCGG ..................((((.(((....))).(((((......))))))))).. ( -9.33 = -8.62 + -0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:50 2011