
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,487,845 – 20,487,947 |
| Length | 102 |
| Max. P | 0.698214 |

| Location | 20,487,845 – 20,487,947 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.02 |
| Shannon entropy | 0.65326 |
| G+C content | 0.49160 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -11.43 |
| Energy contribution | -11.07 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.698214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
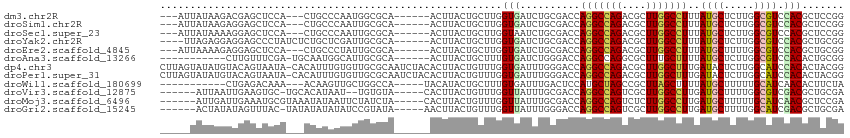
>dm3.chr2R 20487845 102 - 21146708 ---AUUAUAAGACGAGCUCCA---CUGCCCAAUGGCGCA------ACUUACUGCUUGGUGAUCUGCGACCAGGCCAGACGCUUGGCCUUUAUGCUCUUGGCGUCCACGCUCCGG ---..........((((....---..(((...(((((((------..(((((....)))))..)))).))))))..((((((.(((......)))...))))))...))))... ( -33.00, z-score = -1.31, R) >droSim1.chr2R 18949423 102 - 19596830 ---AUUAUAAGAGGAGCUCCA---CUGCCCAAUUGCGCA------ACUUACUGCUUGGUGAUCUGCGACCAGGCCAGACGCUUGGCCUUUAUGCUCUUGGCGUCCACGCUCCGG ---.........(((((....---..(((......((((------..(((((....)))))..))))....)))..((((((.(((......)))...))))))...))))).. ( -33.40, z-score = -1.28, R) >droSec1.super_23 361369 102 - 989336 ---AUUAUAAAAGGAGCUCCA---CUGCCCAAUUGCGCA------ACUUACUGCUUGGUAAUCUGCGACCAGGCCAGACGCUUGGCCUUUAUGCUCUUGGCGUCCACGCUCCGG ---.........(((((....---..(((......((((------..(((((....)))))..))))....)))..((((((.(((......)))...))))))...))))).. ( -33.30, z-score = -1.89, R) >droYak2.chr2R 20459636 104 - 21139217 ----UUAGAGGAGGAGCCCUAUCUCUGCUCGAUUGCGCA------ACUUACUGCUUGGUGAUCUGCGACCAGGCCAGACGCUUGGCCUUUAUGCUCUUGGCGUCCACGCUGCGG ----...(((.((((......)).)).))).....((((------.(.....(((((((........)))))))..((((((.(((......)))...))))))...).)))). ( -32.90, z-score = 0.31, R) >droEre2.scaffold_4845 21862774 102 - 22589142 ---AUUAAAAGAGGAGCUCCA---CUGCCCUAUUGCGCA------ACUUACUGCUUGGUGAUCUGCGACCAGGCCAGACGCUUGGCCUUUAUGCUUUUGGCGUCCACGCUGCGG ---........(((.((....---..)))))....((((------.(.....(((((((........)))))))..((((((.(((......)))...))))))...).)))). ( -32.00, z-score = -0.60, R) >droAna3.scaffold_13266 11154779 96 + 19884421 -----------CUUGUUUCGA-UGCAAUGGCAUUGCGCA------ACUUACUGCUUUGUGAUCUGGGACCAGGCCAGGCGCUUUGCUUUUAUGCUCUUGGCGUCCACACUGCGG -----------.((((..(((-(((....)))))).)))------)....((((..((((..(((....)))((((((.((...........)).))))))...))))..)))) ( -32.30, z-score = -1.49, R) >dp4.chr3 2566400 113 + 19779522 CUUAGUAUAUGUACAGUAAUA-CACAUUUGUGUUGCGCAAUCUACACUUACUGUUUGGUGAUUUGGGACCAGGCCAGACGCUUGGCUUUGAUACUCUUGGCAUCCACACUACGG ..(((((..((((..((((((-((....))))))))......))))..)))))..(((((...((((..(((((.....)))))(((..(.....)..))).)))))))))... ( -28.20, z-score = -0.62, R) >droPer1.super_31 577113 113 - 935084 CUUAGUAUAUGUACAGUAAUA-CACAUUUGUGUUGCGCAAUCUACACUUACUGUUUGGUGAUUUGGGACCAGGCCAGACGCUUGGCUUUGAUACUCUUGGCAUCCACACUACGG ..(((((..((((..((((((-((....))))))))......))))..)))))..(((((...((((..(((((.....)))))(((..(.....)..))).)))))))))... ( -28.20, z-score = -0.62, R) >droWil1.scaffold_180699 586532 95 + 2593675 -----------CUGAGACAAA---ACAAGUUGCUGGCCA-----UACAUACUGCUUUGUGAUUUGACUCCAUGCUAGCCGCUUAGCUUUUAUGCUUUUUGCAUCAACACUUCUA -----------.(((..((((---(.((((.((((((..-----((((........))))...((....)).)))))).))))(((......))))))))..)))......... ( -19.40, z-score = -0.58, R) >droVir3.scaffold_12875 5952670 100 + 20611582 ------AUUAAUUGAAGUGC-UGCACAUAAU--UGUGUA-----CACUUACUGUUUGGUUAUUUGCGACCAGGCCAGUCGCUUGGCCUUGAUGCUUUUGGCGUCGACGCUGCGA ------........(((((.-((((((....--))))))-----)))))(((((((((((......))))))).))))(((..(((.(((((((.....))))))).)))))). ( -37.10, z-score = -3.44, R) >droMoj3.scaffold_6496 16731086 103 + 26866924 ------AUUGAUUGAAAUGCGUAAAUAUAAUUCUAUCUA-----CACUUACUGUUUGGUUAUUUGCGACCAGGCCAGUCUCUUGGCCUUGAUGCUUUUUGCAUCAACGCUCCGA ------...(((((......(((.(((......))).))-----).......((((((((......)))))))))))))((..(((.(((((((.....))))))).)))..)) ( -26.80, z-score = -2.85, R) >droGri2.scaffold_15245 3653052 102 + 18325388 ------ACUAUAUAGUUUAC-UAUAUAUAUAUCCGUAUA-----AACUUACUGUUUGGUUAUUUGGGACCAGGCCAGUCGCUUGGCCUUGAUGCUUUUGGCAUCGACGCUGCGA ------..(((((((....)-))))))............-----.....(((((((((((......))))))).))))(((..(((.(((((((.....))))))).)))))). ( -30.80, z-score = -2.75, R) >consensus _____UAUAAGAUGAGCUCCA___CUAUCCUAUUGCGCA______ACUUACUGCUUGGUGAUCUGCGACCAGGCCAGACGCUUGGCCUUUAUGCUCUUGGCGUCCACGCUGCGG .........................................................(((..........(((((((....)))))))..((((.....)))).)))....... (-11.43 = -11.07 + -0.35)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:45 2011