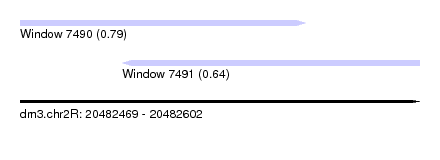
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,482,469 – 20,482,602 |
| Length | 133 |
| Max. P | 0.788253 |

| Location | 20,482,469 – 20,482,564 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.55 |
| Shannon entropy | 0.33350 |
| G+C content | 0.53743 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -15.51 |
| Energy contribution | -17.32 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
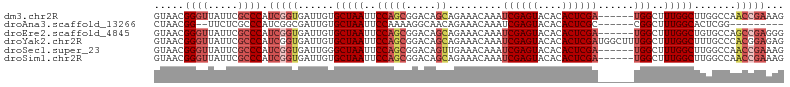
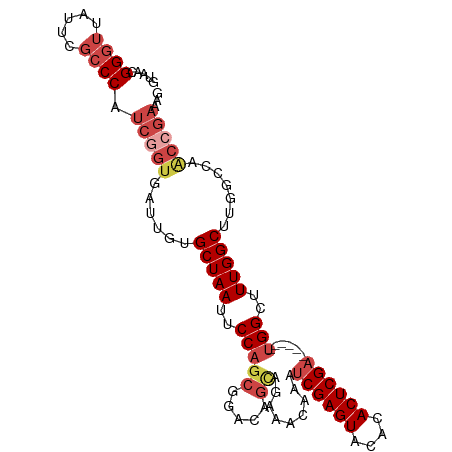
>dm3.chr2R 20482469 95 + 21146708 CCGUCAUUACCUCACCUU------UCUUCCGCGGAGUUGGCUUUCGGUUGGCCAAGCCAAAGCCA------UCGAGUGUGUACUCGAUUUGUUUCUGCUGUCCGCUG ..................------......(((((..(((((((.((((.....)))))))))))------((((((....)))))).............))))).. ( -28.90, z-score = -2.22, R) >droAna3.scaffold_13266 16034901 87 + 19884421 CCGACAUUACCUUACCAC---------CUCACCUUCCUAGCAAUCCG-----AGUGCCAAAGCCG------GCGAGUGUGUACUCGAUUUGUUUCUGUUGCCUUUUG .(((((............---------...........(((((..((-----(((((((..(...------.)...)).)))))))..)))))..)))))....... ( -16.11, z-score = -0.75, R) >droEre2.scaffold_4845 21857376 101 + 22589142 CCGUCAUUACCUCACCUUCCGCGUUCUUCCGCGGAGUUGUCCCUCGGCUGGCACAGCCAAAGCCA------UCGAGUGUGUACUCGAUUUGUUUCUGCUGUCCGCUG ................(((((((......)))))))........((((.((.(((((..((((.(------((((((....)))))))..))))..))))))))))) ( -34.20, z-score = -3.51, R) >droYak2.chr2R 20454287 101 + 21139217 CCGUCAUUACCUCACCUU------UCUUCCGCGGAGUUGGCUCUCCGUGGGCAAAGCCAAAGCCAAAGCCAUCGAGUGUGUACUCGAUUUGUUUCUGCUGUCCGCUG ..................------...(((((((((......)))))))))...(((...(((..((((.(((((((....)))))))..))))..)))....))). ( -31.00, z-score = -2.24, R) >droSec1.super_23 355997 95 + 989336 CCGUCAUUACCUCACCUU------UCUUCCGCGGAGUUGGCUUUCGGUUGGCCAAGCCAAAGCCA------UCGAGUGUGUACUCGAUUUGUUUCAACUGUCCGCUG ..................------......(((((..(((((((.((((.....)))))))))))------((((((....)))))).............))))).. ( -28.90, z-score = -2.49, R) >droSim1.chr2R 18943333 95 + 19596830 CCGUCAUUACCUCACCUU------UCUUCCGCGGAGUUGGCUUUCGGUUGGCCAAGCCAAAGCCA------UCGAGUGUGUACUCGAUUUGUUUCUGCUGUCCGCUG ..................------......(((((..(((((((.((((.....)))))))))))------((((((....)))))).............))))).. ( -28.90, z-score = -2.22, R) >consensus CCGUCAUUACCUCACCUU______UCUUCCGCGGAGUUGGCUUUCGGUUGGCAAAGCCAAAGCCA______UCGAGUGUGUACUCGAUUUGUUUCUGCUGUCCGCUG ..............................(((((..(((((...((((.....))))..)))))......((((((....)))))).............))))).. (-15.51 = -17.32 + 1.81)

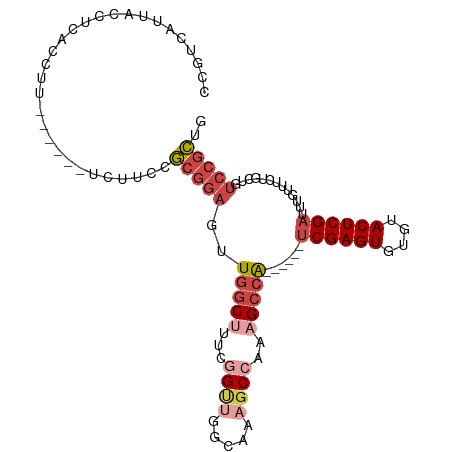
| Location | 20,482,503 – 20,482,602 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 85.81 |
| Shannon entropy | 0.26874 |
| G+C content | 0.51459 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -21.11 |
| Energy contribution | -22.89 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20482503 99 - 21146708 GUAACGGGUUAUUCGCCCAUCGGUGAUUGUGCUAAUUCCAGCGGACAGCAGAAACAAAUCGAGUACACACUCGA------UGGCUUUGGCUUGGCCAACCGAAAG .....((((.....)))).(((((......(((((..((.((.....))........(((((((....))))))------)))..))))).......)))))... ( -31.02, z-score = -1.88, R) >droAna3.scaffold_13266 16034936 88 - 19884421 CUAACGG--UUCUCGCCCAUCGGCGAUUGUGCUAAUUCCAAAAGGCAACAGAAACAAAUCGAGUACACACUCGC------CGGCUUUGGCACUCGG--------- ....(((--...(((((....)))))..(((((((..((....(((....((......))((((....))))))------)))..)))))))))).--------- ( -26.80, z-score = -2.62, R) >droEre2.scaffold_4845 21857416 99 - 22589142 GUAACGGGUUAUUCGCCCAUCGGUGAUUGUGCUAAUUCCAGCGGACAGCAGAAACAAAUCGAGUACACACUCGA------UGGCUUUGGCUGUGCCAGCCGAGGG .....((((.....)))).(((((......(((......)))(((((((.(((....(((((((....))))))------)...))).))))).)).)))))... ( -33.90, z-score = -1.60, R) >droYak2.chr2R 20454321 105 - 21139217 GUAACGGGUUAUUCGCCCAUCGGUGAUUGUGCUAAUUCCAGCGGACAGCAGAAACAAAUCGAGUACACACUCGAUGGCUUUGGCUUUGGCUUUGCCCACGGAGAG .....((((.....)))).((.(((.((.((((...(((...))).)))).))....(((((((....)))))))(((...(((....)))..)))))).))... ( -31.50, z-score = -0.91, R) >droSec1.super_23 356031 99 - 989336 GUAACGGGUUAUUCGCCCAUCGGUGAUUGGGCUAAUUCCAGCGGACAGUUGAAACAAAUCGAGUACACACUCGA------UGGCUUUGGCUUGGCCAACCGAAAG .....((((.....)))).(((((..(..((((((..(((((.....))).......(((((((....))))))------)))..))))))..)...)))))... ( -33.90, z-score = -2.54, R) >droSim1.chr2R 18943367 99 - 19596830 GUAACGGGUUAUUCGCCCAUCGGUGAUUGUGCUAAUUCCAGCGGACAGCAGAAACAAAUCGAGUACACACUCGA------UGGCUUUGGCUUGGCCAACCGAAAG .....((((.....)))).(((((......(((((..((.((.....))........(((((((....))))))------)))..))))).......)))))... ( -31.02, z-score = -1.88, R) >consensus GUAACGGGUUAUUCGCCCAUCGGUGAUUGUGCUAAUUCCAGCGGACAGCAGAAACAAAUCGAGUACACACUCGA______UGGCUUUGGCUUGGCCAACCGAAAG .....((((.....)))).(((((......(((((..(((((.....)).........((((((....))))))......)))..))))).......)))))... (-21.11 = -22.89 + 1.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:44 2011