| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,427,085 – 20,427,205 |
| Length | 120 |
| Max. P | 0.500000 |

| Location | 20,427,085 – 20,427,205 |
|---|---|
| Length | 120 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.72 |
| Shannon entropy | 0.34808 |
| G+C content | 0.47887 |
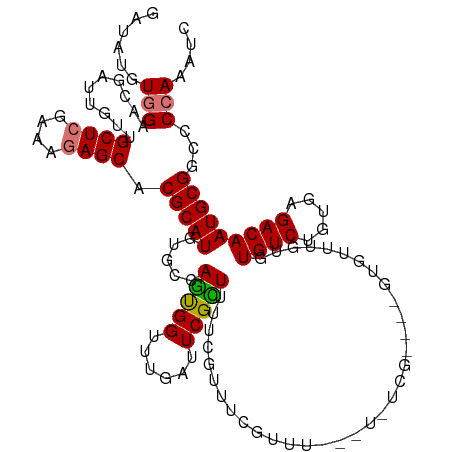
| Mean single sequence MFE | -34.91 |
| Consensus MFE | -21.48 |
| Energy contribution | -21.40 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
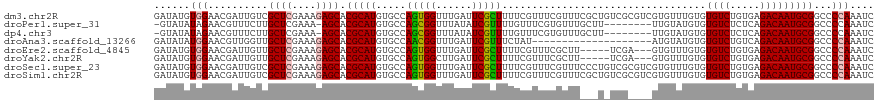
>dm3.chr2R 20427085 120 - 21146708 GAUAUGUGGAACGAUUGUCGCUCGAAAGAGCACGCAUGUGCCAGUGGUUUGAUUCGCUUUUCGUUUCGUUUCGCUGUCGCGUCGUGUUUGUGUGUCUGUGAGACAAUGCGGCCCCAAAUC (((.((.((..((((((((((((....))))(((((((((.((((((..(((..((.....))..)))..)))))).))))).))))..............)))))).))..)))).))) ( -44.70, z-score = -2.83, R) >droPer1.super_31 513874 110 - 935084 -GUAUAUAGAACGUUUCUUGCUCGAAA-AGCACGCAUGUGCCAGCGGUUUAUAUCGUUUUGUUUCGUGUUUGCUU--------UUGUAUGUGUGUCUCUCAGACAAUGCGGCCCCAAAUC -..................(((..(((-((((.(((((.((.((((((....))))))..))..))))).)))))--------))((((((.((.....)).)).)))))))........ ( -24.50, z-score = -0.21, R) >dp4.chr3 2502790 110 + 19779522 -GUAUAUAGAACGUUUCUUGCUCGAAA-AGCACGCAUGUGCCAGCGGUUUAUAUCGUUUUGUUUCGUGUUUGCUU--------UUGUAUGUGUGUCUCUCAGACAAUGCGGCCCCAAAUC -..................(((..(((-((((.(((((.((.((((((....))))))..))..))))).)))))--------))((((((.((.....)).)).)))))))........ ( -24.50, z-score = -0.21, R) >droAna3.scaffold_13266 15977733 100 - 19884421 GAUAUAUGGAACGUUGGUUGCUCGAAAGAGCACGCAUGUGCCAACGGUUUGAUUCGUUUCUAU--------------------AUGUAUGUGUGUCUGUCAGACAAUGCGGCCCCAAAUC .((((((((((((((((((((((....))))).......)))))))(.......)..))))))--------------------)))).((((((((.....))).))))).......... ( -29.61, z-score = -1.40, R) >droEre2.scaffold_4845 21798908 112 - 22589142 GAUAUGUGGAACGAUUGUUGCUCGAAAGAGCACGCAUGUGCCAGUGGUUUGAUUCGCUUUUCGUUUCGCUU-----UCGA---GUGUUUGUGUGUCUGUGAGACAAUGCGGCCCCAAAUC (((.((.((..((((((((((((....))))(((((.(..(((((((..(((........)))..))))).-----..).---)..).)))))........)))))).))..)))).))) ( -33.70, z-score = -1.02, R) >droYak2.chr2R 20396332 112 - 21139217 GAUAUGUGGAACGAUUGUUGCUCGAAAGAGCACGCAUGUGCCAGUGGCUUGAUUCGCUUUUCGUUUCGCUU-----UCGA---GUGUUUGUGUGUCUGUGAGACAAUGCGGCCCCAAAUC (((.((.((..((((((((((((....)))).((((.(..((((..((((((...((..........))..-----))))---))..))).)..).)))).)))))).))..)))).))) ( -36.50, z-score = -1.64, R) >droSec1.super_23 301178 120 - 989336 GAUAUGUGGAACGAUUGUCGCUCGAAAGAGCACGCAUGUGCCAGUGGUUUGAUUCGCUUUUCGUUUCGUUUCCCUGUCGCGUCGUGUUUGUGUGUCUGUGAGACAAUGCGGCCCCAAAUC (((.((.((..((((((((((((....))))(((((((((.(((.((..(((..((.....))..)))...))))).))))).))))..............)))))).))..)))).))) ( -41.10, z-score = -2.41, R) >droSim1.chr2R 18887484 120 - 19596830 GAUAUGUGGAACGAUUGUCGCUCGAAAGAGCACGCAUGUGCCAGUGGUUUGAUUCGCUUUUCGUUUCGUUUCGCUGUCGCGUCGUGUUUGUGUGUCUGUGAGACAAUGCGGCCCCAAAUC (((.((.((..((((((((((((....))))(((((((((.((((((..(((..((.....))..)))..)))))).))))).))))..............)))))).))..)))).))) ( -44.70, z-score = -2.83, R) >consensus GAUAUGUGGAACGAUUGUUGCUCGAAAGAGCACGCAUGUGCCAGUGGUUUGAUUCGCUUUUCGUUUCGUUU___U_UCG____GUGUUUGUGUGUCUGUGAGACAAUGCGGCCCCAAAUC ......(((..........((((....)))).(((((.....(((((......)))))..................................((((.....)))))))))...))).... (-21.48 = -21.40 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:43 2011