| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,413,577 – 20,413,683 |
| Length | 106 |
| Max. P | 0.998699 |

| Location | 20,413,577 – 20,413,683 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 63.32 |
| Shannon entropy | 0.71329 |
| G+C content | 0.40883 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -9.15 |
| Energy contribution | -10.45 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.564255 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
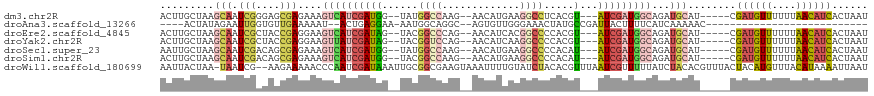
>dm3.chr2R 20413577 106 + 21146708 AUUAGUGAUGUUAAAAAACAUCG-----AUGCAUCUGCCAUCGAU---ACGUGAGGCCUUCAUGUU--CUUGGCCAUA--CCAUCGAUGACUUUCUCGCUCCCGAUUGCUUAGCAAGU .....(((((((....)))))))-----.(((......(((((((---..(((.((((........--...)))).))--).)))))))........((........))...)))... ( -27.60, z-score = -1.72, R) >droAna3.scaffold_13266 15961342 82 + 19884421 ---------------------------GUUUUUGAUGAAAAGUAAUCGGCAUAGUUUCCCAACACU--GCCUGCCAUU-UUCCUCAGU--AUUUUUCAACACCAAUUCUAUAGU---- ---------------------------.....((.(((((((((...((((((((........)))--)..))))...-........)--)))))))).)).............---- ( -11.04, z-score = -1.00, R) >droEre2.scaffold_4845 21782592 106 + 22589142 AUUAGUGAUGUUAAAAAACAUCG-----AUGCAUCUGCCAUCGAU---ACGUGGGGCCGUGAUGUU--CUGGGCCGUA--CUAUCGAUGACUUCCUCGGUAGCGAUUGCUUAGCAAGU ....((((((((....)))))).-----..(((..((((((((((---(.(((.((((..(.....--)..)))).))--)))))))))(((.....))).)))..)))...)).... ( -33.40, z-score = -1.58, R) >droYak2.chr2R 20382094 106 + 21139217 AUUAGUGAUGUUAAAAAACAUCG-----AUGCAUCUGCCAUCGAU---ACGUGGGGCCUUGAUGUU--CUGGACCGUA--CUAUCGAUAACUUCCUCGGUAGCGAUUGCUUAGCAAGU ....((((((((....)))))).-----..(((.(((((((((((---(.(((.((((........--..)).)).))--)))))))).........)))))....)))...)).... ( -27.50, z-score = -0.73, R) >droSec1.super_23 287813 106 + 989336 AUUAGUGAUGUUAAAAAACAUCG-----AUGCAUCUGCCAUCGAU---AUGUGGGGCCUUCAUGUU--CUUGGCCAUA--CCAUCGAUGACUUUCUCGCUGUCGAUUGCUUAGCAAUU .....(((((((....)))))))-----.(((....((.((((((---(.((((((...((((...--..(((.....--)))...))))...))))))))))))).))...)))... ( -29.00, z-score = -1.64, R) >droSim1.chr2R 18874051 106 + 19596830 AUUAGUGAUGUUAAAAAACAUCG-----AUGCAUCUGCCAUCGAU---AUGUGGGGCCUUCAUGUU--CUUGGCCGUA--CCAUCGAUGACUUUCUCGCUGUCGAUUGCUUAGCAAGU .....(((((((....)))))))-----.(((......(((((((---..(((.((((........--...)))).))--).)))))))........((........))...)))... ( -29.70, z-score = -1.44, R) >droWil1.scaffold_180699 2032133 115 + 2593675 AUUAAUUUUAUGUAAACAUGUAGUAAACGUGUAGAUAAAAACGAUUAAACGUGUAGAUACAAAAUUUACUUCGCCGCAAUUUAUCGAUUGGGUUUUUCUU--CGAUUA-UUAGUAAUU (((((((((((.(..((((((.....))))))).)))))).(((..(((...((((((.....))))))...(((.(((((....)))))))).)))..)--))....-))))).... ( -16.80, z-score = 0.69, R) >consensus AUUAGUGAUGUUAAAAAACAUCG_____AUGCAUCUGCCAUCGAU___ACGUGGGGCCUUCAUGUU__CUUGGCCGUA__CCAUCGAUGACUUUCUCGCUACCGAUUGCUUAGCAAGU ......((((((....))))))................(((((((......((.((((.............)))).))....)))))))............................. ( -9.15 = -10.45 + 1.30)


| Location | 20,413,577 – 20,413,683 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 63.32 |
| Shannon entropy | 0.71329 |
| G+C content | 0.40883 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -12.31 |
| Energy contribution | -14.08 |
| Covariance contribution | 1.77 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.998699 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
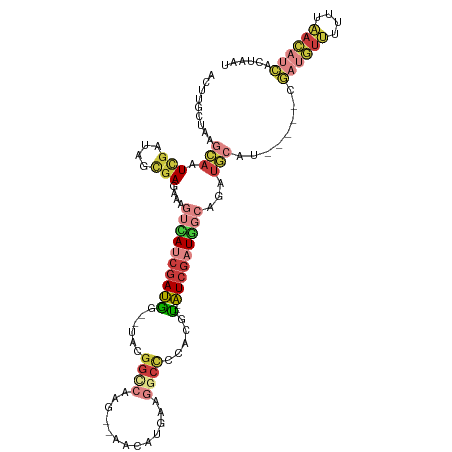
>dm3.chr2R 20413577 106 - 21146708 ACUUGCUAAGCAAUCGGGAGCGAGAAAGUCAUCGAUGG--UAUGGCCAAG--AACAUGAAGGCCUCACGU---AUCGAUGGCAGAUGCAU-----CGAUGUUUUUUAACAUCACUAAU .........(((.(((....)))....(((((((((((--(..((((...--........))))..)).)---)))))))))...)))..-----.((((((....))))))...... ( -34.60, z-score = -3.12, R) >droAna3.scaffold_13266 15961342 82 - 19884421 ----ACUAUAGAAUUGGUGUUGAAAAAU--ACUGAGGAA-AAUGGCAGGC--AGUGUUGGGAAACUAUGCCGAUUACUUUUCAUCAAAAAC--------------------------- ----......((.(..(((((....)))--))..)((((-(.(((..(((--(....(((....)))))))..))).))))).))......--------------------------- ( -19.80, z-score = -2.60, R) >droEre2.scaffold_4845 21782592 106 - 22589142 ACUUGCUAAGCAAUCGCUACCGAGGAAGUCAUCGAUAG--UACGGCCCAG--AACAUCACGGCCCCACGU---AUCGAUGGCAGAUGCAU-----CGAUGUUUUUUAACAUCACUAAU .((((...(((....)))..))))...(((((((((((--(..((((..(--.....)..))))..)).)---)))))))))........-----.((((((....))))))...... ( -32.30, z-score = -3.21, R) >droYak2.chr2R 20382094 106 - 21139217 ACUUGCUAAGCAAUCGCUACCGAGGAAGUUAUCGAUAG--UACGGUCCAG--AACAUCAAGGCCCCACGU---AUCGAUGGCAGAUGCAU-----CGAUGUUUUUUAACAUCACUAAU .((((...(((....)))..))))...(((((((((((--(..((((..(--.....)..))))..)).)---)))))))))........-----.((((((....))))))...... ( -28.40, z-score = -2.17, R) >droSec1.super_23 287813 106 - 989336 AAUUGCUAAGCAAUCGACAGCGAGAAAGUCAUCGAUGG--UAUGGCCAAG--AACAUGAAGGCCCCACAU---AUCGAUGGCAGAUGCAU-----CGAUGUUUUUUAACAUCACUAAU .........(((.(((....)))....(((((((((((--(..((((...--........))))..)).)---)))))))))...)))..-----.((((((....))))))...... ( -32.10, z-score = -3.16, R) >droSim1.chr2R 18874051 106 - 19596830 ACUUGCUAAGCAAUCGACAGCGAGAAAGUCAUCGAUGG--UACGGCCAAG--AACAUGAAGGCCCCACAU---AUCGAUGGCAGAUGCAU-----CGAUGUUUUUUAACAUCACUAAU .((((((...(....)..))))))...(((((((((((--(..((((...--........))))..)).)---)))))))))........-----.((((((....))))))...... ( -33.40, z-score = -3.64, R) >droWil1.scaffold_180699 2032133 115 - 2593675 AAUUACUAA-UAAUCG--AAGAAAAACCCAAUCGAUAAAUUGCGGCGAAGUAAAUUUUGUAUCUACACGUUUAAUCGUUUUUAUCUACACGUUUACUACAUGUUUACAUAAAAUUAAU .........-......--........((((((......)))).))....((((((..((((..((.((((.((............)).)))).)).)))).))))))........... ( -10.50, z-score = 1.40, R) >consensus ACUUGCUAAGCAAUCGAUAGCGAGAAAGUCAUCGAUGG__UACGGCCAAG__AACAUGAAGGCCCCACGU___AUCGAUGGCAGAUGCAU_____CGAUGUUUUUUAACAUCACUAAU .........(((.(((....)))....(((((((((.......((((.............)))).........)))))))))...)))........((((((....))))))...... (-12.31 = -14.08 + 1.77)
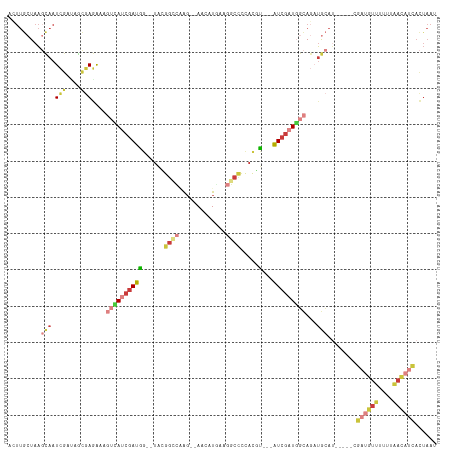
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:42 2011