| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,412,817 – 20,412,920 |
| Length | 103 |
| Max. P | 0.500000 |

| Location | 20,412,817 – 20,412,920 |
|---|---|
| Length | 103 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 73.00 |
| Shannon entropy | 0.57571 |
| G+C content | 0.67174 |
| Mean single sequence MFE | -49.85 |
| Consensus MFE | -14.99 |
| Energy contribution | -14.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 20412817 103 - 21146708 ACGCUG------GCAGC---GGCGCACCACCAGCAGCAGCUGCACCACCACGCCGCUGCAGCUGGUCACCUGAGCCA---CGUCGGCGGUGGGCACGCAGCCAGUAACCACCUGG ..((((------((.((---(((.((((((((((.(((((.((........)).))))).)))))).......(((.---....))))))).)).))).)))))).......... ( -54.50, z-score = -2.70, R) >droSim1.chr2R 18873292 103 - 19596830 ACGCUG------GCAGC---GGCGCACCACCAGCAGCAGCUGCACCACCACGCCGCUGCAGCUGGUCACCUGAGCCA---CGUCGGCGGUGGCCACGCAGCCAGUAACCACCUGG ..((((------((.((---(((.((((((((((.(((((.((........)).))))).)))))).......(((.---....))))))))))..)).)))))).......... ( -50.90, z-score = -1.98, R) >droSec1.super_23 287053 103 - 989336 ACGCUG------GCAGC---GGCGCACCACCAGCAGCAGCUGCACCACCACGCCGCUGCAGCUGGUCACCUGAGCCA---CGUCGGCGGUGGCCACGCAGCCAGUAACCACCUGG ..((((------((.((---(((.((((((((((.(((((.((........)).))))).)))))).......(((.---....))))))))))..)).)))))).......... ( -50.90, z-score = -1.98, R) >droYak2.chr2R 20381321 103 - 21139217 ACGCUG------GCAGC---GGCGCACCACCAGCAGCAGCUGCACCACCACGCCGCUGCCGCUGGCCACCUGAGCCA---CGUCGGCGGCGGCCACGCAGCCAGUAACCACCUGG ..((((------(((((---((((......((((....))))........))))))))))(((((((.((...(((.---....))))).))))).)))))(((.......))). ( -49.44, z-score = -1.43, R) >droEre2.scaffold_4845 21781842 103 - 22589142 ACGCUG------GCAGC---GGCGCACCAUCAGCAGCAGCUGCACCACCACGCCGCCGCUGCCGGGCACCUGAGCCA---CGUCGGCGGCGGCCACGCAGCCAGUAACCACCUGG ..((((------((.((---(((((..........)).)))))........((((((((((.(((((......))).---)).))))))))))......)))))).......... ( -50.90, z-score = -1.83, R) >droAna3.scaffold_13266 15960626 103 - 19884421 ACGCUG------GCCGC---CGCGCACCACCAGCAGCAGCUGCAUCACCAUGCCGCCGCCGCUGGCCACCUGGGUCA---CGUAGGUGGUGGUCAUGCGGCCAGCAAUCAUCUGG ..((((------(((((---.((.((((((((((.((.((.((((....)))).)).)).)))((((.....)))).---....)))))))))...))))))))).......... ( -54.00, z-score = -2.57, R) >dp4.chr3 1523620 100 + 19779522 ACGCUG------GCCGC---UGCACAUCAUCAGCAGCAGUUGCACCAUCA---CGCUGCCGCCGGCCACCUGGGCCA---UGUCGGCGGUGGACACGCGGCCAGCAAUCAUUUGG ..((((------(((((---(((.........)))))....((....((.---(((((((((.((((.....)))).---.).))))))))))...))))))))).......... ( -48.40, z-score = -2.68, R) >droPer1.super_34 718487 100 + 916997 ACGCUG------GCCGC---UGCACAUCAUCAGCAGCAGUUGCACCAUCA---CGCUGCCGCCGGCCACCUGGGCCA---UGUCGGCGGUGGACACGCGGCCAGCAAUCAUUUGG ..((((------(((((---(((.........)))))....((....((.---(((((((((.((((.....)))).---.).))))))))))...))))))))).......... ( -48.40, z-score = -2.68, R) >droWil1.scaffold_180699 2031424 109 - 2593675 ACUUUGGCGGCGGCGGCCCAUGCCCAUCAUCAGCAACAAUUGCACCAUCA---UGCCGCCGCUGGACAUUUAGGACA---UGUGGGUGGCGGACAUGCCGCAAGUAACCAUUUGG ..((..(((((((((((....)))........(((.....))).......---.))))))))..)).........((---.((((...((((.....))))......)))).)). ( -40.80, z-score = -0.73, R) >droMoj3.scaffold_6496 11107844 112 - 26866924 ACGCUGGCUGCCGCUGC---UGCCCAUCAUCAGCAGCAGCUGCACCAUCAUGCUGCUGCAACUGGGCACCUGAGCCAUGGCGUUGGCGGCGGCCAUGCGGCUAGCAAUCAUUUGG ..(((((((((.(((((---((((...(..((((((((((((......)).)))))))).....(((......))).))..)..)))))))))...))))))))).......... ( -56.30, z-score = -1.89, R) >droGri2.scaffold_15245 1889893 112 + 18325388 ACGCUGGCCGCCGCCGC---UGCCCAUCAUCAGCAGCAGCUGCAUCAUCAUGCCGCCGCCACCGGCCAUCUCAGUCAUGGCGUUGGCGGCGGCCAUGCGGCCAGCAAUCAUUUGG ..(((((((((.((.((---(((............))))).))........((((((((((.((.((((.......)))))).))))))))))...))))))))).......... ( -61.30, z-score = -4.63, R) >droVir3.scaffold_12875 3716672 112 + 20611582 ACGCUGGCCGCCGCCGC---GGCCCAUCAUCAGCAGCAGCUGCAUCAUCAUGCCGCGGCCACCGGCCAUUUGAGCCAUGGCGUUGGCGGCGGCCAUGCGGCCAGCAAUCAUUUGG .((((((((((((((((---(((..((.((((((....)))).)).))...))))).((((..((((....).))).))))...))))))))))).))).((((.......)))) ( -58.30, z-score = -2.33, R) >anoGam1.chr2L 10785080 98 + 48795086 ------------GCUGGC--CGUGUUCAAUC-UCAGCGUGUACACCACCGAGGCGGAACUGUACGAUACGUUUUCCA-AGUUUGGUCCACUGCGA-AAGACGACCGUGGUGCUGG ------------(((((.--...........-))))).....((((((...((.(((((.(((...)))))))))).-.....((((.....(..-..)..)))))))))).... ( -23.90, z-score = 1.97, R) >consensus ACGCUG______GCAGC___GGCGCACCAUCAGCAGCAGCUGCACCACCACGCCGCUGCCGCUGGCCACCUGAGCCA___CGUCGGCGGCGGCCACGCGGCCAGCAACCAUCUGG ..((((......((.......)).......)))).((....)).(((.......((.......(((.......)))........(((.(((....))).))).)).......))) (-14.99 = -14.77 + -0.22)

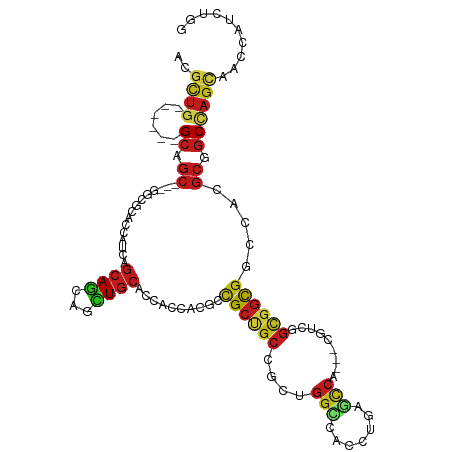

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:40 2011