
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,389,136 – 20,389,314 |
| Length | 178 |
| Max. P | 0.877145 |

| Location | 20,389,136 – 20,389,314 |
|---|---|
| Length | 178 |
| Sequences | 5 |
| Columns | 195 |
| Reading direction | forward |
| Mean pairwise identity | 77.32 |
| Shannon entropy | 0.39162 |
| G+C content | 0.45050 |
| Mean single sequence MFE | -57.66 |
| Consensus MFE | -29.54 |
| Energy contribution | -30.66 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
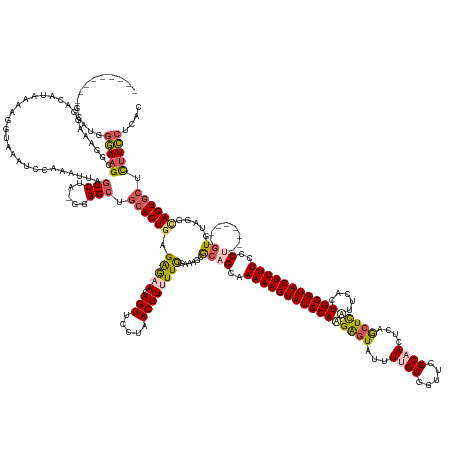
>dm3.chr2R 20389136 178 + 21146708 ---------GCAUGGGAAGCGGAAAGGACAUAAAAGGUAAAUCCAAAUUUGCCUAGGGGGAUGCACUGAGAGAACAGUUCUUAACUUUUUCGAAAGGGCAGGAAAAAGUAUGCAUGAGUAUUUUGUGGUUUGCAACUCAGCUCAUUCACUGCAUACUUUUCCCU--------GUGUAGGCAGUGCUCUUCCUCAC ---------((.......))..............((((((((....))))))))(((((((.((((((.(((((.(((.....))))))))....(.(((((.((((((((((((((((...((((.....))))....))))).....))))))))))).)))--------)).)...)))))))))))))... ( -56.80, z-score = -2.04, R) >droEre2.scaffold_4845 21756386 182 + 22589142 ---GCAAUGGAAGCACAAGCAGCAAAGCUAUAAAAGCUAAUCUCAAGUC-GCCUG-GGGGCUGCACUGAGGGAAUAGUUCCCAACUUCUUUGCUGGCACAGCAAAAAGUAUGCAAGGGUAUUUUGUGGUUCGCUACCCAGCUCCUUCACUGCAUACUUUUCCCC--------GUGCAGCGAGUGCUUCUGCUCAC ---(((...(((((((..(((((..((((.....))))..(((((....-...))-)))))))).....((((.....)))).......((((((.(((.(..(((((((((((((((....(((.(((.....))))))..))))...)))))))))))..).--------))))))))))))))))))).... ( -65.60, z-score = -2.27, R) >droYak2.chr2R 20357634 192 + 21139217 GCAACGGAGGCAAUGGAAGC--AAAGGGCAUGAAAGCUAAAUCCAAAUCAGCCUA-GUGGCUGCACUGAGGAAAUAGUUCUGCACUUCUUUGAAAGCACAGCAAAAAGUAUGCAAGAGUGUUUUGUGGUGUGCCACCCAAGUUAUUCACUGCAUACUUUUUCCCUUUUCUCAGUGUACUUAGUACUUUUCCUCAC ......((((.((.((..((--.....))......(((((.........((((..-..))))((((((((((((..(((.(((............))).)))((((((((((((.(((((.((((.((((...)))))))).)))))..))))))))))))...))))))))))))..))))).)).)))))).. ( -59.50, z-score = -1.98, R) >droSec1.super_23 261845 178 + 989336 ---------GCAUGGGAAGCGGAAAGGACAUAAAAGGUAACUCCAAAUUAGCCUAAGGGGCUUCACUGAGAGAACAGUUGCUAACUUUUUCGAAAGGGAAGCAAAAAGUAUGCAAGAGUAUUUUGUGGUUCGCAACUCUGCUCUCUCAUUGCAUACUUUUCCCU--------GUGUAGACAGUGCUCUUCCUCAC ---------....((((((.(.(..(.((((...((((............)))).(((((((((.((..(((((.(((.....))))))))...)).))))).(((((((((((((((((..(((((...)))))...)))))).....)))))))))))))))--------))))...)..).).))))))... ( -51.50, z-score = -1.02, R) >droSim1.chr2R 18848118 178 + 19596830 ---------GCAUGGGAAGCGGAAAGGACAUAAAAGGUAAAUCCAAAUUAGCCCAAGGGGCUGCACUGAGAGAACAGUUCCUAACUUUUUCGAAUGAGCAGCAAAAAGUAUGCAAGAGUAUUUUGUGGUUCGCAACUCUGCUCUCUCAUUGCAUACUUUUUCCU--------GUGUAGGCAGUGCUCUUCCUCAC ---------....((((((.(((....((.......))...))).....(((((...)))))((((((.(((((.(((.....))))))))......((((.((((((((((((((((((..(((((...)))))...)))))).....)))))))))))).))--------)).....)))))).))))))... ( -54.90, z-score = -1.67, R) >consensus _________GCAUGGGAAGCGGAAAGGACAUAAAAGGUAAAUCCAAAUUAGCCUA_GGGGCUGCACUGAGAGAACAGUUCCUAACUUUUUCGAAAGCGCAGCAAAAAGUAUGCAAGAGUAUUUUGUGGUUCGCAACUCAGCUCAUUCACUGCAUACUUUUCCCU________GUGUAGGCAGUGCUCUUCCUCAC ..............(((((((......((((....((.....))............((((((((.((..(((((.(((.....))))))))...)).))))).((((((((((((((((...((((.....))))....))))).....)))))))))))))).........))))......))))..))).... (-29.54 = -30.66 + 1.12)

| Location | 20,389,136 – 20,389,314 |
|---|---|
| Length | 178 |
| Sequences | 5 |
| Columns | 195 |
| Reading direction | reverse |
| Mean pairwise identity | 77.32 |
| Shannon entropy | 0.39162 |
| G+C content | 0.45050 |
| Mean single sequence MFE | -56.20 |
| Consensus MFE | -31.64 |
| Energy contribution | -31.60 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20389136 178 - 21146708 GUGAGGAAGAGCACUGCCUACAC--------AGGGAAAAGUAUGCAGUGAAUGAGCUGAGUUGCAAACCACAAAAUACUCAUGCAUACUUUUUCCUGCCCUUUCGAAAAAGUUAAGAACUGUUCUCUCAGUGCAUCCCCCUAGGCAAAUUUGGAUUUACCUUUUAUGUCCUUUCCGCUUCCCAUGC--------- (((.(((((.....((((((..(--------(((((((((((((((((......))(((((...............)))))))))))))))))))))..................(.((((......)))).).......)))))).....((((...........))))......))))))))..--------- ( -48.86, z-score = -2.26, R) >droEre2.scaffold_4845 21756386 182 - 22589142 GUGAGCAGAAGCACUCGCUGCAC--------GGGGAAAAGUAUGCAGUGAAGGAGCUGGGUAGCGAACCACAAAAUACCCUUGCAUACUUUUUGCUGUGCCAGCAAAGAAGUUGGGAACUAUUCCCUCAGUGCAGCCCC-CAGGC-GACUUGAGAUUAGCUUUUAUAGCUUUGCUGCUUGUGCUUCCAUUGC--- (..((((.(((((((((((((((--------((.((((((((((((((......)))(((((.............)))))..))))))))))).))))).))))....(((((((((.....))))........(((..-..)))-))))))))...((((.....))))....))))).))))..).....--- ( -69.72, z-score = -2.79, R) >droYak2.chr2R 20357634 192 - 21139217 GUGAGGAAAAGUACUAAGUACACUGAGAAAAGGGAAAAAGUAUGCAGUGAAUAACUUGGGUGGCACACCACAAAACACUCUUGCAUACUUUUUGCUGUGCUUUCAAAGAAGUGCAGAACUAUUUCCUCAGUGCAGCCAC-UAGGCUGAUUUGGAUUUAGCUUUCAUGCCCUUU--GCUUCCAUUGCCUCCGUUGC (.((((..(((((((((((.(((((((.(((.(((((((((((((((.((.....(((((((...)))).))).....))))))))))))))).(((..((((....))))..)))..)).))).)))))))(((((..-..))))))))))).....((......))....)--))))......)))))..... ( -61.50, z-score = -2.39, R) >droSec1.super_23 261845 178 - 989336 GUGAGGAAGAGCACUGUCUACAC--------AGGGAAAAGUAUGCAAUGAGAGAGCAGAGUUGCGAACCACAAAAUACUCUUGCAUACUUUUUGCUUCCCUUUCGAAAAAGUUAGCAACUGUUCUCUCAGUGAAGCCCCUUAGGCUAAUUUGGAGUUACCUUUUAUGUCCUUUCCGCUUCCCAUGC--------- (((.(((((....((((....))--------))(((((.(((((((.(((((((((((((((((((..............))))).)))..(((((...((((....))))..)))))))))))))))).)).((((.....)))).................)))).).))))).))))))))..--------- ( -49.14, z-score = -1.34, R) >droSim1.chr2R 18848118 178 - 19596830 GUGAGGAAGAGCACUGCCUACAC--------AGGAAAAAGUAUGCAAUGAGAGAGCAGAGUUGCGAACCACAAAAUACUCUUGCAUACUUUUUGCUGCUCAUUCGAAAAAGUUAGGAACUGUUCUCUCAGUGCAGCCCCUUGGGCUAAUUUGGAUUUACCUUUUAUGUCCUUUCCGCUUCCCAUGC--------- (((.(((((.((((((......(--------((.(((((((((((((.(((...((......))(.....)......)))))))))))))))).))).......(((..(((.....))).)))...))))))(((((...))))).....((((...........))))......))))))))..--------- ( -51.80, z-score = -1.59, R) >consensus GUGAGGAAGAGCACUGCCUACAC________AGGGAAAAGUAUGCAGUGAAAGAGCUGAGUUGCGAACCACAAAAUACUCUUGCAUACUUUUUGCUGCCCUUUCGAAAAAGUUAGGAACUGUUCUCUCAGUGCAGCCCC_UAGGCUAAUUUGGAUUUACCUUUUAUGUCCUUUCCGCUUCCCAUGC_________ (((.(((((.((((((...............((.(((((((((((((..........((((...............))))))))))))))))).)).............(((.....))).......))))))((((.....)))).......................)))))))).................. (-31.64 = -31.60 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:38 2011