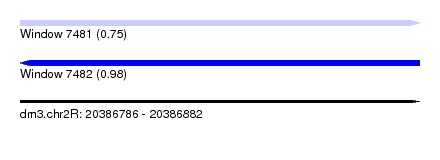
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,386,786 – 20,386,882 |
| Length | 96 |
| Max. P | 0.983624 |

| Location | 20,386,786 – 20,386,882 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 60.64 |
| Shannon entropy | 0.68180 |
| G+C content | 0.50164 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -13.87 |
| Energy contribution | -13.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.754741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
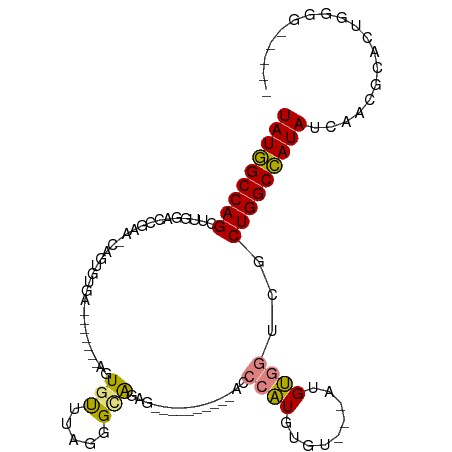
>dm3.chr2R 20386786 96 + 21146708 UAUGGCCAGCGUGGACCGAA-UGGUGUGA-------AGUGUUUAGGGCAGAGAGAGUG---CACCCAUGUGU---UAGUGGUCGCUGGCCAUAUCAACGCAUUGGGGAAA-- ((((((((((...(((((.(-(((.(((.-------(.(.(((.......))).).).---)))))))....---...)))))))))))))))((..(.....)..))..-- ( -30.50, z-score = -0.64, R) >droEre2.scaffold_4845 21754075 90 + 22589142 UAUGGCCAGCUUUGACCAAAACAGUGUGGGCUA---AGUGUUUGGGGCAG--------------CCAUGUGU---AUGUGGUAUCUGGCCAUAUCAACGCACUGGGG--AAU (((((((((...((.((.(((((.(........---).))))).)).))(--------------((((....---..)))))..)))))))))((..(.....)..)--).. ( -26.60, z-score = 0.56, R) >droYak2.chr2R 20355156 111 + 21139217 UAUGGCCAGCUUUGAACAAA-CAGUGUGAACUAGUAAGUGUUUAGGGUAGAGUACCCAAGGUACCCAUAGGUGGUAUGUGGUAUCUGGCCAUAUCAACGCACUGGGGGCAAU (((((((.((((((((((.(-((((....))).))...))))))))))(((.((((((.(.((((....)))).).)).)))))))))))))).....((.(....)))... ( -35.30, z-score = -0.84, R) >droSec1.super_23 259499 88 + 989336 UAUGGCCAGCUUGAACCGAA-UAGUGUGA-------AGUGUUUAGGGCAGAGAGAGCA---CACCCAUGUGU---AAGUGGUCACUGGCCAUAUCAACGCAU---------- (((((((((..(((.(((..-..(..((.-------.((((((..........)))))---)...))..)..---...))))))))))))))).........---------- ( -25.60, z-score = -0.99, R) >droSim1.chr2R 18845880 88 + 19596830 UAUGGCCAGCUUGAACCGAA-UAGUGUGA-------AGUGUUUAGGGCAGAGAGAGCA---CACCCAUGUGU---AAGUGGUCACUGGCCAUAUCAACGCAU---------- (((((((((..(((.(((..-..(..((.-------.((((((..........)))))---)...))..)..---...))))))))))))))).........---------- ( -25.60, z-score = -0.99, R) >dp4.chr3 1498404 87 - 19779522 UAUCGCCAGCGAACUCUUUUGCAGUGUGU--------CUUCAUAGUGUGAG------------UGUGUGAGUGUGGUGCUGGCGCUGGCGAUAUCCAUGCACUGAGA----- ((((((((((..((((....(((.((((.--------...)))).))))))------------)..((.(((.....))).))))))))))))..............----- ( -31.80, z-score = -1.54, R) >droPer1.super_34 690570 87 - 916997 UAUCGCCAGCGAACUCUUUUGCAGUGUGU--------CUUCAUAGUGUGAG------------UGUGUGAGUGUGGUGCUGGCGCUGGCGAUAUCCAUGCACUGAGA----- ((((((((((..((((....(((.((((.--------...)))).))))))------------)..((.(((.....))).))))))))))))..............----- ( -31.80, z-score = -1.54, R) >consensus UAUGGCCAGCUUGGACCGAA_CAGUGUGA_______AGUGUUUAGGGCAGAG__________ACCCAUGUGU___AUGUGGUCGCUGGCCAUAUCAACGCACUGGGG_____ (((((((((.............................(((.....)))...............((((.........))))...)))))))))................... (-13.87 = -13.43 + -0.44)

| Location | 20,386,786 – 20,386,882 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 60.64 |
| Shannon entropy | 0.68180 |
| G+C content | 0.50164 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -14.41 |
| Energy contribution | -14.19 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
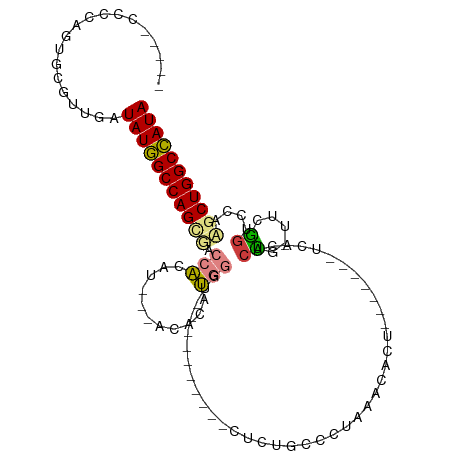
>dm3.chr2R 20386786 96 - 21146708 --UUUCCCCAAUGCGUUGAUAUGGCCAGCGACCACUA---ACACAUGGGUG---CACUCUCUCUGCCCUAAACACU-------UCACACCA-UUCGGUCCACGCUGGCCAUA --.................(((((((((((.......---.....((((.(---((.......)))))))......-------....(((.-...)))...))))))))))) ( -28.90, z-score = -3.22, R) >droEre2.scaffold_4845 21754075 90 - 22589142 AUU--CCCCAGUGCGUUGAUAUGGCCAGAUACCACAU---ACACAUGG--------------CUGCCCCAAACACU---UAGCCCACACUGUUUUGGUCAAAGCUGGCCAUA ...--..............(((((((((..((((...---(((...((--------------(((...........---))))).....)))..)))).....))))))))) ( -23.50, z-score = -0.83, R) >droYak2.chr2R 20355156 111 - 21139217 AUUGCCCCCAGUGCGUUGAUAUGGCCAGAUACCACAUACCACCUAUGGGUACCUUGGGUACUCUACCCUAAACACUUACUAGUUCACACUG-UUUGUUCAAAGCUGGCCAUA ...................(((((((((.((((.((((.....))))))))....(((((...))))).....((..((.(((....))))-)..))......))))))))) ( -26.70, z-score = -0.05, R) >droSec1.super_23 259499 88 - 989336 ----------AUGCGUUGAUAUGGCCAGUGACCACUU---ACACAUGGGUG---UGCUCUCUCUGCCCUAAACACU-------UCACACUA-UUCGGUUCAAGCUGGCCAUA ----------.........((((((((((((((....---((((....)))---)((.......))..........-------........-...))))...)))))))))) ( -24.00, z-score = -1.16, R) >droSim1.chr2R 18845880 88 - 19596830 ----------AUGCGUUGAUAUGGCCAGUGACCACUU---ACACAUGGGUG---UGCUCUCUCUGCCCUAAACACU-------UCACACUA-UUCGGUUCAAGCUGGCCAUA ----------.........((((((((((((((....---((((....)))---)((.......))..........-------........-...))))...)))))))))) ( -24.00, z-score = -1.16, R) >dp4.chr3 1498404 87 + 19779522 -----UCUCAGUGCAUGGAUAUCGCCAGCGCCAGCACCACACUCACACA------------CUCACACUAUGAAG--------ACACACUGCAAAAGAGUUCGCUGGCGAUA -----..............(((((((((((..(((..............------------.(((.....)))..--------.....((.....)).)))))))))))))) ( -19.70, z-score = -0.61, R) >droPer1.super_34 690570 87 + 916997 -----UCUCAGUGCAUGGAUAUCGCCAGCGCCAGCACCACACUCACACA------------CUCACACUAUGAAG--------ACACACUGCAAAAGAGUUCGCUGGCGAUA -----..............(((((((((((..(((..............------------.(((.....)))..--------.....((.....)).)))))))))))))) ( -19.70, z-score = -0.61, R) >consensus _____CCCCAGUGCGUUGAUAUGGCCAGCGACCACAU___ACACAUGGGU__________CUCUGCCCUAAACACU_______UCACACUG_UUCGGUCCAAGCUGGCCAUA ...................(((((((((((.(((...........)))........................................((.....))....))))))))))) (-14.41 = -14.19 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:36 2011