
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,296,514 – 20,296,637 |
| Length | 123 |
| Max. P | 0.752839 |

| Location | 20,296,514 – 20,296,634 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Shannon entropy | 0.49628 |
| G+C content | 0.56652 |
| Mean single sequence MFE | -45.36 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.54 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.66 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.623594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
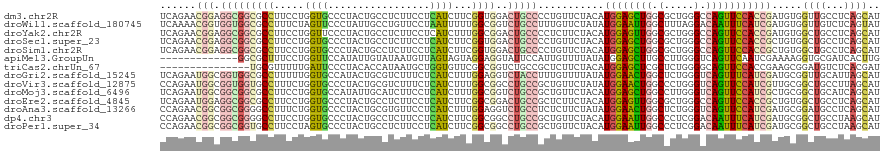
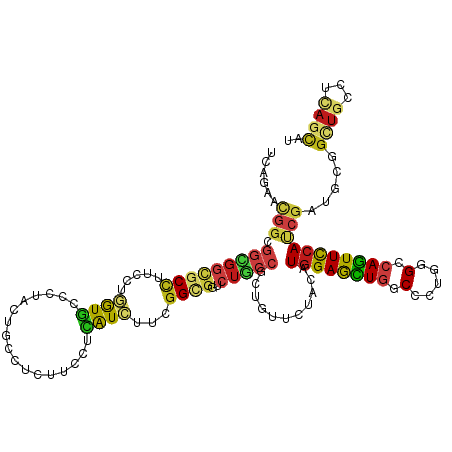
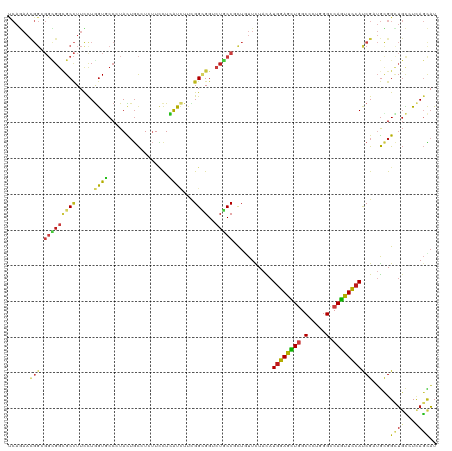
>dm3.chr2R 20296514 120 - 21146708 UCAGAACGGAGGCGGCGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGUGGACUGCCCCUGUUCUACAUGGAGCUGGCGCUGGGCCAGUUCCACCGAUGUGGUUGCCUCAGCAU ........(((((((((((.....))))))............((.((((....((((((.......).))))).((((((((((.....))))))))))..)))).))..)))))..... ( -50.20, z-score = -1.72, R) >droWil1.scaffold_180745 1430420 120 - 2843958 UCAAAACGGUGGUGGCGCCUUUCUAGUUCCCUAUUGCCUGUUCCUAAUUUUUGGCGGUCUGCCUUUGUUCUAUAUGGAAUUGGCUUUAGGACAAUUUCAUCGAUGUGGUUGUCUCAGUAU .......((((....))))...........(((..(((.(((((........(((.....)))............))))).)))..)))((((((..((....))..))))))....... ( -28.25, z-score = -0.19, R) >droYak2.chr2R 20258644 120 - 21139217 UCAGAACGGAGGCGGCGCCUUCCUGGUUCCCUACUGCCUCUUCCUCAUCUUUGGCGGACUGCCCCUCUUCUACAUGGAGUUGGCGCUGGGCCAGUUCCACCGAUGUGGCUGCCUCAGCAU ........((((((((......(((((((....(((((..............)))))...((((..((((.....))))..)).)).)))))))...(((....)))))))))))..... ( -42.54, z-score = -0.15, R) >droSec1.super_23 166748 120 - 989336 UCAGAACGGAGGCGGCGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGUGGACUGCCCCUGUUCUACAUGGAGCUGGCGCUGGGCCAGUUCCACCGCUGUGGCUGCCUCAGCAU ..(((((((.(((((((((.....))))))...(..((..............))..)...))).)))))))...((((((((((.....))))))))))..((((.((...)).)))).. ( -55.94, z-score = -2.95, R) >droSim1.chr2R 18749844 120 - 19596830 UCAGAACGGAGGCGGCGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGUGGACUGCCCCUGUUCUACAUGGAGCUGGCGCUGGGCCAGUUCCACCGCUGUGGCUGCCUCAGCAU ..(((((((.(((((((((.....))))))...(..((..............))..)...))).)))))))...((((((((((.....))))))))))..((((.((...)).)))).. ( -55.94, z-score = -2.95, R) >apiMel3.GroupUn 137016520 107 + 399230636 -------------GGCGCUUUCCUGGUUCCAUAUUGUAUAAUGUUAGUAGUAGGAGGUAUUCCAUUGUUUUAUAUGGAGCUUGCCUUGGGUCAGUUCAAUCGAAAAGGUGCGAUCACUUG -------------.(((((((..(((((.(((((...((((((..((((.(....).)))).))))))...)))))(((((.(((...))).))))))))))..)))))))......... ( -25.90, z-score = -0.47, R) >triCas2.chrUn_67 73620 105 - 258350 ---------------UGCGUUUUUGAUUCCCUACACCAUAAUGCUGGUGUUCGGCGGUCUGCCGCUCUUCUACAUGGAGCUCGCUCUGGGGCAGUUCCACCGAAGCGGAUGUCUCACGAU ---------------..(((....((((((((((((((......))))))((((.((.((((((((((.......)))))...(....))))))..)).)))))).))).)))..))).. ( -36.50, z-score = -1.43, R) >droGri2.scaffold_15245 965381 120 - 18325388 UCAGAAUGGCGGUGGCGCCUUUUUGGUGCCAUACUGCGUCUUUCUCAUCUUUGGAGGUCUACCUUUGUUUUAUAUGGAACUGGCUCUGGGUCAGUUUCAUCGAUGCGGUUGCAUUAGCAU ........(((((((((((.....))))))))((((((((............(((((....))))).......(((((((((((.....))))))))))).)))))))))))........ ( -45.00, z-score = -3.84, R) >droVir3.scaffold_12875 7416864 120 - 20611582 CCAGAAUGGCGGUGGUGCCUUUCUGGUGCCCUACUGCGUCUUUCUCAUCUUUGGCGGCCUGCCGCUGUUCUAUAUGGAACUGGCCCUGGGUCAGUUCCAUCGUUGCGGCUGCCUUAGCAU ..((((..(((((((..((.....))..))..)))))....))))...((..(((((((.((.((........((((((((((((...)))))))))))).)).)))))))))..))... ( -50.30, z-score = -3.05, R) >droMoj3.scaffold_6496 26217144 120 - 26866924 UCAGAAUGGCGGCGGCGCCUUCCUGGUGCCAUAUUGCAUCUUCCUCAUCUUUGGCGGUCUGCCGCUGUUCUACAUGGAGCUGGCCUUGGGUCAGUUCCAUCGCUGCGGCUGCAUCAGCAU ..(((((((((((((((((.....))))))............(((((....))).))...)))))))))))..((((((((((((...)))))))))))).((((.........)))).. ( -53.80, z-score = -2.99, R) >droEre2.scaffold_4845 21656370 120 - 22589142 UCAGAAUGGAGGCGGCGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGCGGACUGCCGCUCUUCUACAUGGAGUUGGCGCUGGGCCAGUUCCACCGCUGUGGCUGCCUCAGCAU ..((((.((.(((((((((.....))))))...(((((..............)))))...))).)).))))...((((..((((.....))))..))))..((((.((...)).)))).. ( -48.44, z-score = -0.94, R) >droAna3.scaffold_13266 15833997 120 - 19884421 CCAGAACGGCGGCGGGGCCUUUCUGGUGCCCUACUGCGUGUUCCUCAUCUUUGGAGGUCUGCCUCUCUUCUAUAUGGAACUGGCUCUGGGUCAGUUCCAUCGAUGCGGAUGCCUCAGCAU ((.(((((((((..((((((....)).))))..)))).)))))..((((...(((((....))))).......(((((((((((.....))))))))))).)))).))((((....)))) ( -50.70, z-score = -2.79, R) >dp4.chr3 1401659 120 + 19779522 CCAGAACGGCGGCGGGGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGCGGCCUGCCGCUGUUCUACAUGGAAUUGGCCCUCGGACAAUUUCAUCGAUGCGGCUGCCUAAGCAU ..(((((((((((((((((.....)))......(((((..............)))))))))))))))))))..(((((((((.((...)).))))))))).......(((.....))).. ( -47.34, z-score = -1.80, R) >droPer1.super_34 593068 120 + 916997 CCAGAACGGCGGCGGUGCCUUCCUAGUGCCCUACUGCCUCUUCCUCAUCUUCGGCGGCCUGCCGCUGUUCUACAUGGAAUUGGCCCUCGGACAAUUUCAUCGAUGCGGCUGCCUAAGCAU ..(((((((((((((.(((.....((((...))))(((..............)))))))))))))))))))..(((((((((.((...)).))))))))).......(((.....))).. ( -44.24, z-score = -1.89, R) >consensus UCAGAACGGCGGCGGCGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGCGGUCUGCCGCUGUUCUACAUGGAGCUGGCCCUGGGCCAGUUCCAUCGAUGCGGCUGCCUCAGCAU ......(((.(((((((((.....((((.................))))...))))..)))))...........((((((((.(.....).))))))))))).....((((...)))).. (-17.60 = -17.54 + -0.06)
| Location | 20,296,517 – 20,296,637 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.95 |
| Shannon entropy | 0.49700 |
| G+C content | 0.56775 |
| Mean single sequence MFE | -44.09 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.04 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.64 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.752839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
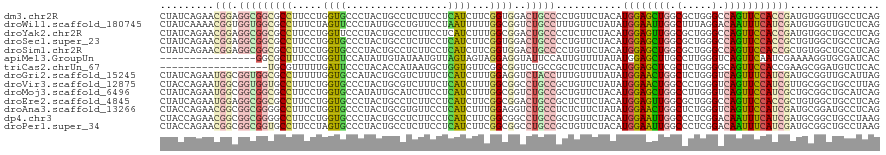
>dm3.chr2R 20296517 120 - 21146708 CUAUCAGAACGGAGGCGGCGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGUGGACUGCCCCUGUUCUACAUGGAGCUGGCGCUGGGCCAGUUCCACCGAUGUGGUUGCCUCAG .(((((((((((.(((((((((.....))))))...(..((..............))..)...))).)))))))...((((((((((.....))))))))))..))))............ ( -50.54, z-score = -2.36, R) >droWil1.scaffold_180745 1430423 120 - 2843958 CUAUCAAAACGGUGGUGGCGCCUUUCUAGUUCCCUAUUGCCUGUUCCUAAUUUUUGGCGGUCUGCCUUUGUUCUAUAUGGAAUUGGCUUUAGGACAAUUUCAUCGAUGUGGUUGUCUCAG ..........((((....))))...........(((..(((.(((((........(((.....)))............))))).)))..)))((((((..((....))..)))))).... ( -28.25, z-score = -0.39, R) >droYak2.chr2R 20258647 120 - 21139217 CUAUCAGAACGGAGGCGGCGCCUUCCUGGUUCCCUACUGCCUCUUCCUCAUCUUUGGCGGACUGCCCCUCUUCUACAUGGAGUUGGCGCUGGGCCAGUUCCACCGAUGUGGCUGCCUCAG ...........((((((((......(((((((....(((((..............)))))...((((..((((.....))))..)).)).)))))))...(((....))))))))))).. ( -42.54, z-score = -0.66, R) >droSec1.super_23 166751 120 - 989336 CUAUCAGAACGGAGGCGGCGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGUGGACUGCCCCUGUUCUACAUGGAGCUGGCGCUGGGCCAGUUCCACCGCUGUGGCUGCCUCAG .....(((((((.(((((((((.....))))))...(..((..............))..)...))).)))))))...((((((((((.....))))))))))...(((.((...)).))) ( -50.74, z-score = -2.05, R) >droSim1.chr2R 18749847 120 - 19596830 CUAUCAGAACGGAGGCGGCGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGUGGACUGCCCCUGUUCUACAUGGAGCUGGCGCUGGGCCAGUUCCACCGCUGUGGCUGCCUCAG .....(((((((.(((((((((.....))))))...(..((..............))..)...))).)))))))...((((((((((.....))))))))))...(((.((...)).))) ( -50.74, z-score = -2.05, R) >apiMel3.GroupUn 137016523 104 + 399230636 ----------------GGCGCUUUCCUGGUUCCAUAUUGUAUAAUGUUAGUAGUAGGAGGUAUUCCAUUGUUUUAUAUGGAGCUUGCCUUGGGUCAGUUCAAUCGAAAAGGUGCGAUCAC ----------------.(((((((..(((((.(((((...((((((..((((.(....).)))).))))))...)))))(((((.(((...))).))))))))))..)))))))...... ( -25.90, z-score = -0.75, R) >triCas2.chrUn_67 73623 102 - 258350 ------------------UGCGUUUUUGAUUCCCUACACCAUAAUGCUGGUGUUCGGCGGUCUGCCGCUCUUCUACAUGGAGCUCGCUCUGGGGCAGUUCCACCGAAGCGGAUGUCUCAC ------------------.........((((((((((((((......))))))((((.((.((((((((((.......)))))...(....))))))..)).)))))).))).))).... ( -34.80, z-score = -1.40, R) >droGri2.scaffold_15245 965384 120 - 18325388 CUAUCAGAAUGGCGGUGGCGCCUUUUUGGUGCCAUACUGCGUCUUUCUCAUCUUUGGAGGUCUACCUUUGUUUUAUAUGGAACUGGCUCUGGGUCAGUUUCAUCGAUGCGGUUGCAUUAG ...........(((((((((((.....))))))))((((((((............(((((....))))).......(((((((((((.....))))))))))).)))))))))))..... ( -45.00, z-score = -4.40, R) >droVir3.scaffold_12875 7416867 120 - 20611582 CUACCAGAAUGGCGGUGGUGCCUUUCUGGUGCCCUACUGCGUCUUUCUCAUCUUUGGCGGCCUGCCGCUGUUCUAUAUGGAACUGGCCCUGGGUCAGUUCCAUCGUUGCGGCUGCCUUAG ...(((((((((((((((..((.....))..))..)))))(.....).))).))))).(((..(((((........((((((((((((...))))))))))))....))))).))).... ( -50.60, z-score = -3.72, R) >droMoj3.scaffold_6496 26217147 120 - 26866924 CUAUCAGAAUGGCGGCGGCGCCUUCCUGGUGCCAUAUUGCAUCUUCCUCAUCUUUGGCGGUCUGCCGCUGUUCUACAUGGAGCUGGCCUUGGGUCAGUUCCAUCGCUGCGGCUGCAUCAG .....(((((((((((((((((.....))))))............(((((....))).))...)))))))))))..((((((((((((...)))))))))))).(.(((....))).).. ( -52.00, z-score = -3.08, R) >droEre2.scaffold_4845 21656373 120 - 22589142 CUAUCAGAAUGGAGGCGGCGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGCGGACUGCCGCUCUUCUACAUGGAGUUGGCGCUGGGCCAGUUCCACCGCUGUGGCUGCCUCAG ...........(((((((((((.....))))))...(((((..............)))))...(((((.........((((..((((.....))))..)))).....))))).))))).. ( -46.98, z-score = -1.05, R) >droAna3.scaffold_13266 15834000 120 - 19884421 CUACCAGAACGGCGGCGGGGCCUUUCUGGUGCCCUACUGCGUGUUCCUCAUCUUUGGAGGUCUGCCUCUCUUCUAUAUGGAACUGGCUCUGGGUCAGUUCCAUCGAUGCGGAUGCCUCAG .((((((((.(((......))).)))))))).....(((.(.(((((.((((...(((((....))))).......(((((((((((.....))))))))))).)))).))).))).))) ( -50.10, z-score = -3.10, R) >dp4.chr3 1401662 120 + 19779522 CUACCAGAACGGCGGCGGGGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGCGGCCUGCCGCUGUUCUACAUGGAAUUGGCCCUCGGACAAUUUCAUCGAUGCGGCUGCCUAAG ....(((...(((((..((((((....)).))))..)))))....((.((((..((((((....))))))......(((((((((.((...)).))))))))).)))).)))))...... ( -46.20, z-score = -2.14, R) >droPer1.super_34 593071 120 + 916997 CUACCAGAACGGCGGCGGUGCCUUCCUAGUGCCCUACUGCCUCUUCCUCAUCUUCGGCGGCCUGCCGCUGUUCUACAUGGAAUUGGCCCUCGGACAAUUUCAUCGAUGCGGCUGCCUAAG ...(((((((((((((((.(((.....((((...))))(((..............)))))))))))))))))))..(((((((((.((...)).)))))))))......))......... ( -42.84, z-score = -2.11, R) >consensus CUAUCAGAACGGCGGCGGCGCCUUCCUGGUGCCCUACUGCCUCUUCCUCAUCUUCGGCGGUCUGCCGCUGUUCUACAUGGAGCUGGCCCUGGGCCAGUUCCAUCGAUGCGGCUGCCUCAG .........(((.(((((((((.....((((.................))))...))))..)))))...........((((((((.(.....).)))))))))))............... (-17.35 = -17.04 + -0.31)
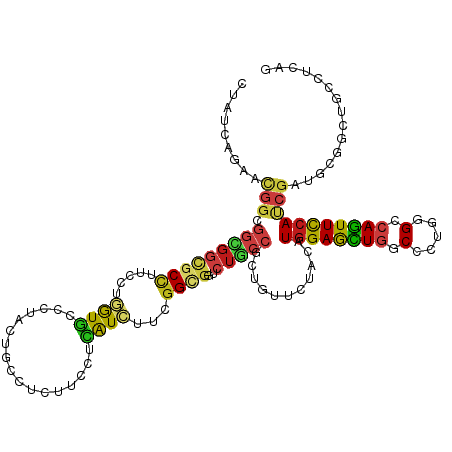
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:22 2011