| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,286,632 – 20,286,733 |
| Length | 101 |
| Max. P | 0.950779 |

| Location | 20,286,632 – 20,286,733 |
|---|---|
| Length | 101 |
| Sequences | 14 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.81 |
| Shannon entropy | 0.54194 |
| G+C content | 0.50606 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.95 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
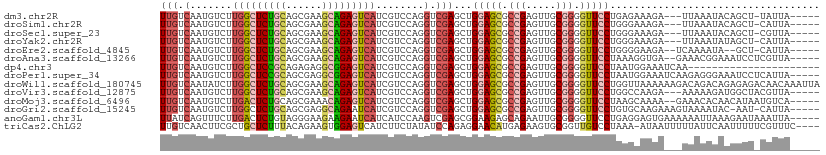
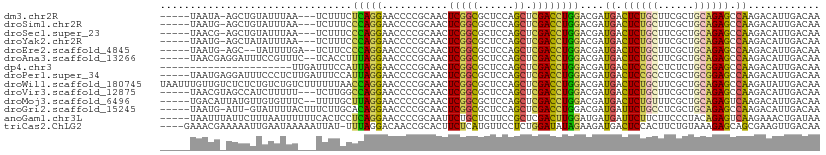
>dm3.chr2R 20286632 101 + 21146708 UUGUCAAUGUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGAGAAAGA---UUAAAUACAGCU-UAUUA----- ......(((...(((((((((......))))))))).))).......((((((((((.(((.....))).))))((.....)).---.......)))))-)....----- ( -32.40, z-score = -0.79, R) >droSim1.chr2R 18738599 101 + 19596830 UUGUCAAUGUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGGGAAAGA---UUAAAUACAGCU-CAUUA----- ......(((...(((((((((......))))))))).))).......((((((((((.(((.....))).))))((.....)).---.......)))))-)....----- ( -35.00, z-score = -1.22, R) >droSec1.super_23 156912 101 + 989336 UUGUCAAUGUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGGGAAAGA---UUAAAUACAGCU-CGUUA----- ......(((...(((((((((......))))))))).)))......(((((((((((.(((.....))).))))((.....)).---.......)))))-))...----- ( -36.10, z-score = -1.36, R) >droYak2.chr2R 20248763 101 + 21139217 UUGUCAAUGUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGGGAAAGA---UUAAAUAUAGCU-CAUUA----- ......(((...(((((((((......))))))))).))).......((((((((((.(((.....))).)))))(((......---))).....))))-)....----- ( -33.30, z-score = -0.88, R) >droEre2.scaffold_4845 21646496 100 + 22589142 UUGUCAAUGUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGGGGAAGA--UCAAAAUA--GCU-CAUUA----- ......(((...(((((((((......))))))))).))).......((((((((((.(((.....))).)))))(((......--)))....)--)))-)....----- ( -35.40, z-score = -1.12, R) >droAna3.scaffold_13266 5750226 103 - 19884421 UUGUCAAUGUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUAAAGGUGA--GAAACGGAAAUCCUCGUUA----- .....((((...(((((((((......)))))))))...(((...((..((((((((.(((.....))).)))))....)))..--))...)))......)))).----- ( -34.30, z-score = -0.57, R) >dp4.chr3 1391233 88 - 19779522 UUGUCAAUGUCUUGGCUCCGCAGAGAGGCGGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUAAUGGAAAUCAA---------------------- ............(((((((((......)))))))))...((((((.....))(((((.(((.....))).)))))...))))......---------------------- ( -32.60, z-score = -1.26, R) >droPer1.super_34 582480 105 - 916997 UUGUCAAUGUCUUGGCUCCGCAGCGAGGCGGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUAAUGGAAAUCAAGAGGGAAAUCCUCAUUA----- .....(((((((((((((((((((..((((((....)).(((((......))))).))))..))))))))))(((....)))...)))))(((....))))))).----- ( -43.00, z-score = -2.58, R) >droWil1.scaffold_180745 1393511 110 + 2843958 UUGUCAAUAUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGGUUAAAAAAGACAGACAGAGAGACAACAAAUUA (((((.......(((((((((......)))))))))..(((...(((.(((((((((.(((.....))).))))).))))......))).))).....)))))....... ( -40.60, z-score = -3.14, R) >droVir3.scaffold_12875 7403965 102 + 20611582 UUGUCAAUGUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGGCCAAGA---AAAAAGAUGGCUACGUUA----- .....(((((..(((((((((......)))))))))(((((....((..((((((((.(((.....))).))))).)))...))---.....)))))..))))).----- ( -36.90, z-score = -1.13, R) >droMoj3.scaffold_6496 26198397 103 + 26866924 UUGUCAAUGUCUUGACUCUGCAGCGAAACAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUAAGCAAAA--GAAACACAACAUAAUGUCA----- ((((.....(((((((((((........))))))).(((.......)))((((((((.(((.....))).)))))..)))..))--))...))))..........----- ( -30.00, z-score = -0.86, R) >droGri2.scaffold_15245 953152 103 + 18325388 UUGUCAAUGUCUUGGCUCUGCAGCGAGGCAGAAUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGUGCAAGAAAGUAAAAUAC-AAU-CAUUA----- .........(((((((((((((((..(((.((....)).(((((......)))))..)))..)))))))))).......)))))...........-...-.....----- ( -31.31, z-score = -0.63, R) >anoGam1.chr3L 4899608 105 + 41284009 UUAUCAGUUUCUUGACUCUGUAGGGAAGAAGAAUCAUCAUCCAAGUCGAGCGGAAGAGCAGAAUUGCGGGGUUCCUGAGGAGUGAAAAAAUUAAAGAAUAAAUUA----- ...(((.(((((((((((((((((((.((.......)).))).....(..(....)..)....))))))))))...)))))))))....................----- ( -22.00, z-score = -0.18, R) >triCas2.ChLG2 7524203 105 + 12900155 UUGUCAACUUCGCUGCUCUUUACAGAAGUGGAGUCAUCUUCUAUAUCCAGAGGAACAUGAGAAGUGCGGUUGUCCUAAA-AUAAUUUUUAUUCAAUUUUUCGUUUC---- ..(.(((((.((((.((((((((....)))))....((((((......))))))....))).)))).))))).).....-..........................---- ( -20.10, z-score = -0.32, R) >consensus UUGUCAAUGUCUUGGCUCUGCAGCGAAGCAGAGUCAUCGUCCAGGUCGAGCUGGAGCGCCGAGUUGCGGGGUUCCUGAGGAAGA___AAAAAUAAAGCU_CAUUA_____ (((.(.......(((((((((......)))))))))........).)))...(((((.(((.....))).)))))................................... (-23.58 = -23.95 + 0.36)
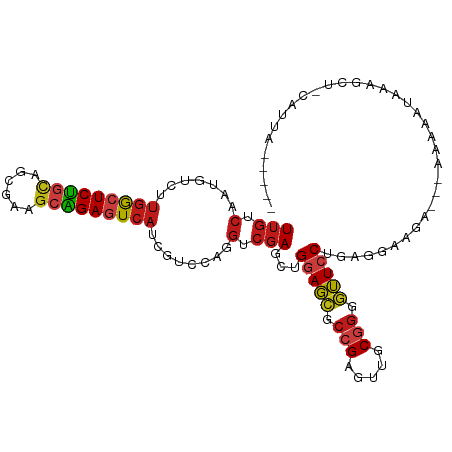
| Location | 20,286,632 – 20,286,733 |
|---|---|
| Length | 101 |
| Sequences | 14 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.81 |
| Shannon entropy | 0.54194 |
| G+C content | 0.50606 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.09 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.61 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
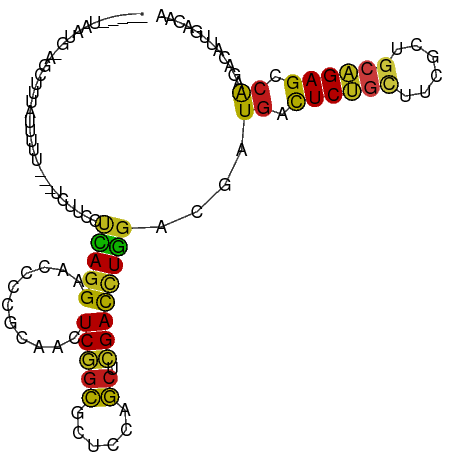
>dm3.chr2R 20286632 101 - 21146708 -----UAAUA-AGCUGUAUUUAA---UCUUUCUCAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCUGCUUCGCUGCAGAGCCAAGACAUUGACAA -----.....-.((((.......---...(((....)))..........))))..(((((......)))))(((((.((((((......))))))......))))).... ( -21.85, z-score = 0.25, R) >droSim1.chr2R 18738599 101 - 19596830 -----UAAUG-AGCUGUAUUUAA---UCUUUCCCAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCUGCUUCGCUGCAGAGCCAAGACAUUGACAA -----...((-(((((.......---...(((....)))...(((......)))...))))))).......(((((.((((((......))))))......))))).... ( -25.20, z-score = -0.68, R) >droSec1.super_23 156912 101 - 989336 -----UAACG-AGCUGUAUUUAA---UCUUUCCCAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCUGCUUCGCUGCAGAGCCAAGACAUUGACAA -----...((-(((((.......---...(((....)))...(((......)))...))))))).......(((((.((((((......))))))......))))).... ( -27.20, z-score = -1.36, R) >droYak2.chr2R 20248763 101 - 21139217 -----UAAUG-AGCUAUAUUUAA---UCUUUCCCAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCUGCUUCGCUGCAGAGCCAAGACAUUGACAA -----(((((-............---......(((((......((......))((....))....)))))....((.((((((......)))))).))...))))).... ( -25.00, z-score = -1.11, R) >droEre2.scaffold_4845 21646496 100 - 22589142 -----UAAUG-AGC--UAUUUUGA--UCUUCCCCAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCUGCUUCGCUGCAGAGCCAAGACAUUGACAA -----(((((-...--........--......(((((......((......))((....))....)))))....((.((((((......)))))).))...))))).... ( -25.00, z-score = -0.76, R) >droAna3.scaffold_13266 5750226 103 + 19884421 -----UAACGAGGAUUUCCGUUUC--UCACCUUUAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCUGCUUCGCUGCAGAGCCAAGACAUUGACAA -----...((((.......(((((--(.......)))))).......)))).((.(((((......))))))).((.((((((......)))))).))............ ( -27.34, z-score = -0.83, R) >dp4.chr3 1391233 88 + 19779522 ----------------------UUGAUUUCCAUUAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCCGCCUCUCUGCGGAGCCAAGACAUUGACAA ----------------------.....((((....))))..(((.....)))((.(((((......))))))).((.((((((......)))))).))............ ( -22.60, z-score = -0.71, R) >droPer1.super_34 582480 105 + 916997 -----UAAUGAGGAUUUCCCUCUUGAUUUCCAUUAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCCGCCUCGCUGCGGAGCCAAGACAUUGACAA -----(((((.((....)).(((((..((((....))))..(((((.(..((((.(((((......))))).((....))))))..).)))))...)))))))))).... ( -29.10, z-score = -1.08, R) >droWil1.scaffold_180745 1393511 110 - 2843958 UAAUUUGUUGUCUCUCUGUCUGUCUUUUUUAACCAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCUGCUUCGCUGCAGAGCCAAGAUAUUGACAA .......(((((....(((((...........(((((......((......))((....))....)))))....((.((((((......)))))).)))))))..))))) ( -29.90, z-score = -1.47, R) >droVir3.scaffold_12875 7403965 102 - 20611582 -----UAACGUAGCCAUCUUUUU---UCUUGGCCAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCUGCUUCGCUGCAGAGCCAAGACAUUGACAA -----..................---(((((((((((......((......))((....))....))))).......((((((......))))))))))))......... ( -28.10, z-score = -0.78, R) >droMoj3.scaffold_6496 26198397 103 - 26866924 -----UGACAUUAUGUUGUGUUUC--UUUUGCUUAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCUGUUUCGCUGCAGAGUCAAGACAUUGACAA -----.........((((((.(((--((......)))))...))))))((((((.(((((......))))))).(((((((((......))))))))).....))))... ( -29.60, z-score = -1.31, R) >droGri2.scaffold_15245 953152 103 - 18325388 -----UAAUG-AUU-GUAUUUUACUUUCUUGCACAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGAUUCUGCCUCGCUGCAGAGCCAAGACAUUGACAA -----(((((-...-.............((((...((....))))))((.((((.(((((......))))))......(((((......)))))))).)).))))).... ( -23.00, z-score = -0.21, R) >anoGam1.chr3L 4899608 105 - 41284009 -----UAAUUUAUUCUUUAAUUUUUUCACUCCUCAGGAACCCCGCAAUUCUGCUCUUCCGCUCGACUUGGAUGAUGAUUCUUCUUCCCUACAGAGUCAAGAAACUGAUAA -----...........................((((.......(((....)))((((..((((.....(((.((.......)).))).....)))).))))..))))... ( -15.50, z-score = -0.09, R) >triCas2.ChLG2 7524203 105 - 12900155 ----GAAACGAAAAAUUGAAUAAAAAUUAU-UUUAGGACAACCGCACUUCUCAUGUUCCUCUGGAUAUAGAAGAUGACUCCACUUCUGUAAAGAGCAGCGAAGUUGACAA ----..........................-....((....))..(((((....(((.((((...((((((((.((....)))))))))).)))).))))))))...... ( -17.80, z-score = -0.31, R) >consensus _____UAAUG_AGCUUUAUUUUU___UCUUCCUCAGGAACCCCGCAACUCGGCGCUCCAGCUCGACCUGGACGAUGACUCUGCUUCGCUGCAGAGCCAAGACAUUGACAA ................................(((((...........(((((......)).))))))))....((.((((((......)))))).))............ (-16.74 = -16.09 + -0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:20 2011