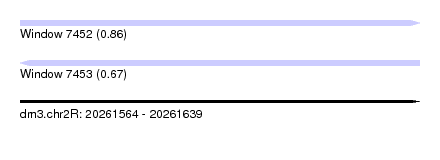
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,261,564 – 20,261,639 |
| Length | 75 |
| Max. P | 0.858067 |

| Location | 20,261,564 – 20,261,639 |
|---|---|
| Length | 75 |
| Sequences | 13 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 84.61 |
| Shannon entropy | 0.37603 |
| G+C content | 0.55982 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -19.99 |
| Energy contribution | -19.65 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858067 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
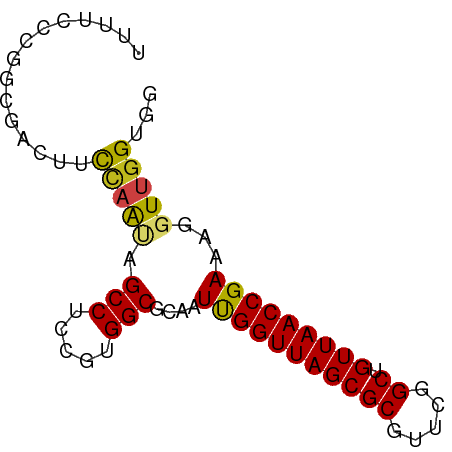
>dm3.chr2R 20261564 75 + 21146708 UUUUCCCGGCGAGUUCCAAUAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG .....((((((.((((((((.(((.....)))...)))))..))).)).))))(..((((((....))))))..) ( -27.90, z-score = -1.70, R) >droSim1.chr2R 18712804 75 + 19596830 UUUUCCCGGCGACUUCCAAUAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG .....((((((.((.(((((.(((.....)))...)))))..)))))...)))(..((((((....))))))..) ( -27.70, z-score = -1.59, R) >droSec1.super_23 132123 75 + 989336 UUUUUCCAGCGAGUUCCAAUAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG .....((((((.((((((((.(((.....)))...)))))..))).)))).))(..((((((....))))))..) ( -26.50, z-score = -1.88, R) >droEre2.scaffold_4845 21619815 75 + 22589142 UUUCCUCGGCGAAUCUCAAUAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG .......(((...........))).(((..((((.........))))...)))(..((((((....))))))..) ( -23.50, z-score = -0.60, R) >droYak2.chr2R 20222253 75 + 21139217 UUUUCCCGUCGAGUUCCAACUGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG .....(((.((.(((((((.((((.....))))...))))..))).))..)))(..((((((....))))))..) ( -25.00, z-score = -1.14, R) >droAna3.scaffold_13266 5724480 75 - 19884421 UUCGCCAGGGCGCUUCCAAGAGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG ..(((((.((.((((....))))..)).)))))...(((((((((((.....)).)))))))))........... ( -29.40, z-score = -1.05, R) >dp4.chr3 1366035 75 - 19779522 UUGUCGGUGCGUUUUUCAAGAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG ..(((((((((((..((((..(((.....)))....))))..)))))).)))))..((((((....))))))... ( -27.40, z-score = -1.85, R) >droPer1.super_34 556152 75 - 916997 UUGUCGGUGCGUUUUUCAAGAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG ..(((((((((((..((((..(((.....)))....))))..)))))).)))))..((((((....))))))... ( -27.40, z-score = -1.85, R) >droWil1.scaffold_180697 1329238 75 - 4168966 UGAAAUUCGAUGGUUUUCGUAGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG ...........((((.....)))).(((..((((.........))))...)))(..((((((....))))))..) ( -22.50, z-score = -0.37, R) >droVir3.scaffold_12875 7374087 75 + 20611582 UUUGCCGCUAGAUUUUCAACCGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG ....(((((((.(((((...((((.....)))).....(((((((((.....)).)))))))))))).))))))) ( -28.60, z-score = -1.87, R) >droMoj3.scaffold_6496 26168815 75 + 26866924 UUUUCCACGCGGCCCUCAGUUGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG ....((((.(((((....((.(((.....)))))..(((((((((((.....)).)))))))))..))))))))) ( -30.60, z-score = -2.01, R) >droGri2.scaffold_15245 924757 74 + 18325388 -UUUUCGGCUGGCCGCCAACUGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG -...........((((((((((((.....)))....(((((((((((.....)).)))))))))..))))))))) ( -30.80, z-score = -1.65, R) >anoGam1.chr2L 9211308 65 + 48795086 ----------AAAUUCAUAUCGCCUUCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG ----------.....(((..((((.....))))...(((((((((((.....)).))))))))).......))). ( -21.10, z-score = -1.51, R) >consensus UUUUCCCGGCGACUUCCAAUAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGG ...............(((((.(((.....)))....(((((((((((.....)).)))))))))...)))))... (-19.99 = -19.65 + -0.34)

| Location | 20,261,564 – 20,261,639 |
|---|---|
| Length | 75 |
| Sequences | 13 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 84.61 |
| Shannon entropy | 0.37603 |
| G+C content | 0.55982 |
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -16.22 |
| Energy contribution | -16.29 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.665860 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
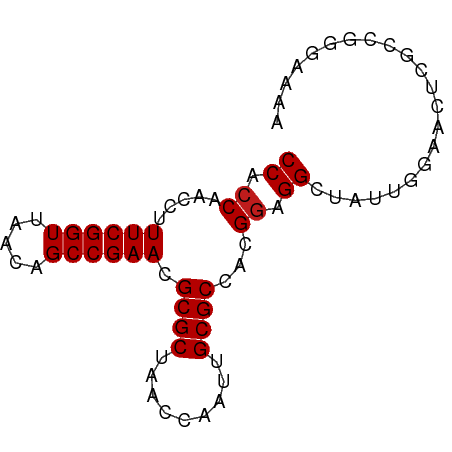
>dm3.chr2R 20261564 75 - 21146708 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUAUUGGAACUCGCCGGGAAAA ...((.....((((((.....)))))).(((....(((((...(((.....))).)))))....))).))..... ( -21.10, z-score = -0.73, R) >droSim1.chr2R 18712804 75 - 19596830 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUAUUGGAAGUCGCCGGGAAAA ...((.....((((((.....)))))).((((.........))))..(((.((((......)))).))))).... ( -23.00, z-score = -0.99, R) >droSec1.super_23 132123 75 - 989336 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUAUUGGAACUCGCUGGAAAAA ...(((....((((((.....)))))).(((....(((((...(((.....))).)))))....))))))..... ( -21.80, z-score = -1.64, R) >droEre2.scaffold_4845 21619815 75 - 22589142 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUAUUGAGAUUCGCCGAGGAAA ...((..((.((((((.....)))))).((((.........))))...)).(((...........)))..))... ( -20.00, z-score = -0.98, R) >droYak2.chr2R 20222253 75 - 21139217 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCAGUUGGAACUCGACGGGAAAA ...((.....((((((.....))))))((((....((((((((.(....)..))))))))....)).)))).... ( -22.90, z-score = -1.35, R) >droAna3.scaffold_13266 5724480 75 + 19884421 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCUCUUGGAAGCGCCCUGGCGAA .(.(((....((((((.....)))))).(((((..((((..(.(((.....))).))))).)))))..))).).. ( -24.20, z-score = -0.75, R) >dp4.chr3 1366035 75 + 19779522 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUCUUGAAAAACGCACCGACAA ..........((((((.....)))))).((((.........))))..(((..((..((....)).)).))).... ( -17.70, z-score = -1.11, R) >droPer1.super_34 556152 75 + 916997 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUCUUGAAAAACGCACCGACAA ..........((((((.....)))))).((((.........))))..(((..((..((....)).)).))).... ( -17.70, z-score = -1.11, R) >droWil1.scaffold_180697 1329238 75 + 4168966 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCUACGAAAACCAUCGAAUUUCA ((.((.....((((((.....)))))).((((.........))))...)).))...(((......)))....... ( -17.90, z-score = -1.39, R) >droVir3.scaffold_12875 7374087 75 - 20611582 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCGGUUGAAAAUCUAGCGGCAAA ....(((((..(((((((...((.....))..)))))))....(((.....))))))))................ ( -21.00, z-score = -0.31, R) >droMoj3.scaffold_6496 26168815 75 - 26866924 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCAACUGAGGGCCGCGUGGAAAA ((((.......(((((((...((.....))..)))))))..(((((.(..((....))..))).))))))).... ( -25.70, z-score = -1.23, R) >droGri2.scaffold_15245 924757 74 - 18325388 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCAGUUGGCGGCCAGCCGAAAA- .........((((((((....((((..........((((((((.(....)..)))))))))))).)))))))).- ( -23.90, z-score = -0.96, R) >anoGam1.chr2L 9211308 65 - 48795086 CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGAAGGCGAUAUGAAUUU---------- ........(..(((((((...((.....))..)))))))..)((((.....))))..........---------- ( -16.20, z-score = -1.74, R) >consensus CCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUAUUGGAACUCGCCGGGAAAA ((.((.....((((((.....)))))).((((.........))))...)).))...................... (-16.22 = -16.29 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:12 2011