| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,846,078 – 3,846,183 |
| Length | 105 |
| Max. P | 0.935745 |

| Location | 3,846,078 – 3,846,183 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 67.88 |
| Shannon entropy | 0.61380 |
| G+C content | 0.44293 |
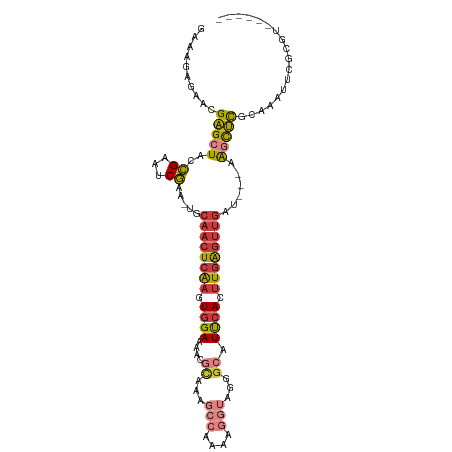
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -11.70 |
| Energy contribution | -12.56 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
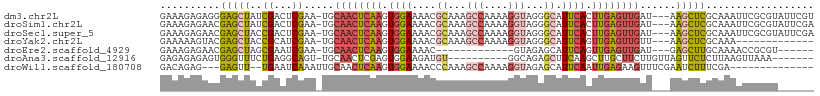
>dm3.chr2L 3846078 105 - 23011544 GAAAGAGAGGGAGCUAUCGACUCGAA-UGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAU---AAGCUCGCAAAUUCGCGUAUUCGU (((.......((((((((((((((((-(((.......(((.....)))...(((....)))...))))))....))))))))---.)))))((......))...))).. ( -32.80, z-score = -2.74, R) >droSim1.chr2L 3801116 105 - 22036055 GAAAGAGAACGAGCUAUCGACUCGAA-UGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAU---AAGCUCGCAAAUUCGCGUAUUCGA ....(((.((((((((((((((((((-(((.......(((.....)))...(((....)))...))))))....))))))))---.)))))((......)))).))).. ( -34.90, z-score = -3.53, R) >droSec1.super_5 1947846 105 - 5866729 GAAAGAGAACGAGCUACCGACUCGAA-UGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAU---AAGCUCGCAAAUUCGCGUAUUCGA ....(((.(((((((..((...))..-..(((((((((((((....((...(((....)))...)).)))))))))))))..---.)))))((......)))).))).. ( -33.00, z-score = -2.98, R) >droYak2.chr2L 3851260 92 - 22324452 GAAAAAGUACGAGCUACCGCAUCGAA-UGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCAGUUGAGUUGUU---AAGCUCGCAAA------------- .........((((((...((((...)-)))(((((((.((((....((...(((....)))...)).)))).)))))))...---.))))))....------------- ( -27.50, z-score = -2.80, R) >droEre2.scaffold_4929 3897806 86 - 26641161 GAAAGAGAACGAGCUAGCGAAUCGAA-UGCAACUCAAGUGGAAAAC-------------GUAGAGCAUUCAGUUGAGUUGAU---GAGCUUGCAAAACCGCGU------ ............((.(((...(((..-..((((((((.((((....-------------........)))).)))))))).)---))))).))..........------ ( -18.50, z-score = 0.05, R) >droAna3.scaffold_12916 1851569 91 - 16180835 GAGAGAGAGUGGGUUUCUGAGGCAGU-UGCAACUCGAGUGGAAGAUGU----------GGCAGAGCUUCAAGCUUGCUUCUUGUUAGUUCUCUUAAGUUAAA------- (((((((((..((((..((((((..(-(((.((((........)).))----------.)))).))))))))))..))).........))))))........------- ( -26.10, z-score = -0.08, R) >droWil1.scaffold_180708 9735121 90 - 12563649 GACAGAG---GAGUU--UGAAUCAAAUUGCAACUCAAGUGGAAAACCCAAAGCCAAAAGGUAGAGCAUUCAAUUGAGAAGUUUCGAAUCUUUCGA-------------- ....(((---..(((--((((((.(((((...(((...(((.....)))..(((....))).)))....)))))..))...)))))))..)))..-------------- ( -16.80, z-score = -0.06, R) >consensus GAAAGAGAACGAGCUACCGAAUCGAA_UGCAACUCAAGUGGAAAACGCAAAGCCAAAAGGUAGGGCAUUCACUUGAGUUGAU___AAGCUCGCAAAUUCGCGU______ ..........(((((..((...)).....((((((((.((((....((...(((....)))...)).)))).))))))))......))))).................. (-11.70 = -12.56 + 0.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:46 2011