| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,251,843 – 20,251,947 |
| Length | 104 |
| Max. P | 0.748315 |

| Location | 20,251,843 – 20,251,947 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 64.18 |
| Shannon entropy | 0.61955 |
| G+C content | 0.45360 |
| Mean single sequence MFE | -24.96 |
| Consensus MFE | -7.93 |
| Energy contribution | -6.67 |
| Covariance contribution | -1.26 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20251843 104 - 21146708 --------------AAUGUGAACUUUAAAGCCACUUAAUAUCCAAUUCUCUGAGCUGUGCCUGCUCGACUGGAUAUAUGGAAUUACCAUAUGGAACUCCCGCUUCGAAGCUCCCACAA --------------..((((..(((..((((......(((((((....((.((((.......)))))).)))))))..(((.((.((....)))).))).))))..)))....)))). ( -26.30, z-score = -3.32, R) >droSim1.chr2R 18702715 103 - 19596830 --------------AAUGUGAACUUC-AAGCCACUAAAUAUCCAAUUCCCUGAGCUGUGCCUGCUCGAGUGGAUGUAUGGAAUUACCAUAUGGAACUCCCGCUUCGAAGCUCCCACAA --------------..((((..((((-((((......(((((((.(((...((((.......))))))))))))))..(((.((.((....)))).))).)))).))))....)))). ( -27.60, z-score = -2.84, R) >droSec1.super_23 122473 103 - 989336 --------------AAUGUGAACUUC-AUGCCACUAAAUAUCCAAUUCCCUGAGCUGUGCCUGCUCGAGUGGAUAUAUGGAAUUUCCAUAUGGAACUCCCGCUUCGAAGCUCCCACAA --------------..((((..((((-..((......(((((((.(((...((((.......))))))))))))))..(((..((((....)))).))).))...))))....)))). ( -26.50, z-score = -2.60, R) >droEre2.scaffold_4845 21609825 102 - 22589142 --------------AAUGUGGAGUUU-GGGCCACUAAAUAUCCGAUUUUCUGGGCUUUGCCCGCUCG-GUAGAUGUGUGCAAUUACCAUAUGGAACCCCCGAUUCAAAGCUCCCACAA --------------..(((((.((((-((....))))))............((((((((.......(-((((.((....)).)))))...(((.....)))...))))))))))))). ( -28.00, z-score = -0.71, R) >droYak2.chr2R 20211982 103 - 21139217 --------------AAUGUGAAGUUU-AGACCACCAAAUAUCCGAUUUUCUGGGCUUAGCCCGCUCGAGUAGAUGUGUGGCAUUACCAUAUGGAACCCCCGAUUCAAAGCUCCCAAAA --------------..((.(.(((((-.((((((((..((..(((.....(((((...))))).)))..))..)).))))..........(((.....)))..)).))))).)))... ( -20.90, z-score = -0.15, R) >dp4.chr3 1354954 103 + 19779522 AGGGAACCAUUUUGUAGUUCAAUUAUUAACCCACUAAAUAUGCAGUUUUCUGGGCUUUUCCUCUACGAGCACACGCGAGCACACA-CGCGUAUAAUUUCCCCCU-------------- .(((((....(((((((............((((..((((.....))))..))))........)))))))...(((((........-))))).....)))))...-------------- ( -22.65, z-score = -1.95, R) >droPer1.super_34 545069 103 + 916997 AGGGAACCAUUUUGUAGUUCAAUUAUUAACCCACUAAAUAUGCAGUUUUCUGGGCUUUUCCUCUACGAGCACACGCGAGCACACA-CGCGUAUAAUUCCCCCCU-------------- .(((((....(((((((............((((..((((.....))))..))))........)))))))...(((((........-)))))....)))))....-------------- ( -22.75, z-score = -1.92, R) >consensus ______________AAUGUGAACUUU_AAGCCACUAAAUAUCCAAUUUUCUGGGCUUUGCCUGCUCGAGUAGAUGUGUGGAAUUACCAUAUGGAACUCCCGCUUCGAAGCUCCCACAA ........................................(((.((((..(((((.......)))))...))))(((((.......))))))))........................ ( -7.93 = -6.67 + -1.26)

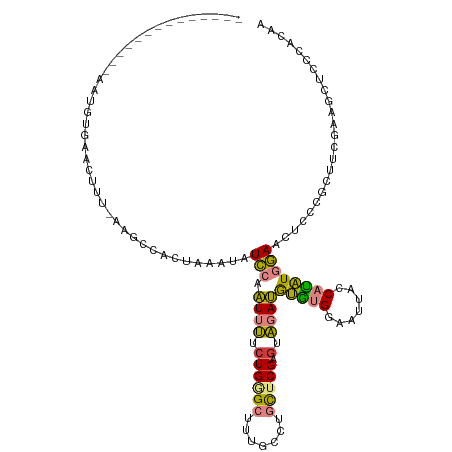

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:09 2011