
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,250,548 – 20,250,652 |
| Length | 104 |
| Max. P | 0.688044 |

| Location | 20,250,548 – 20,250,652 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.45 |
| Shannon entropy | 0.42382 |
| G+C content | 0.52163 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -24.97 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
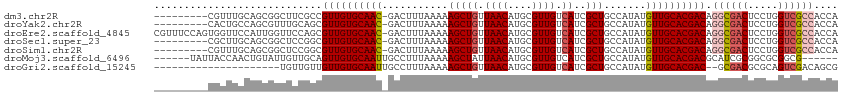
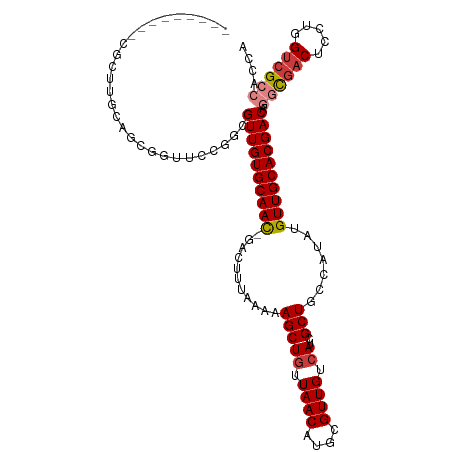
>dm3.chr2R 20250548 104 + 21146708 ---------CGUUUGCAGCGGCUUCGCCGUUGUGCAAC-GACUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACAGGCGACUCCUGGUCGCCACCA ---------........(((((((((((((((((((((-(.........(((((.((((....)))).))..)))......))))))))))).))))).....))))))..... ( -35.66, z-score = -1.21, R) >droYak2.chr2R 20210630 104 + 21139217 ---------CACUGCCAGCGUUUGCAGCGUUGUGCAAC-GACUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACAGGCGACUCCUGGUCGCCACCA ---------...(((.(((((..((((((.((.(((((-(...((((.......))))....)))))))).))))))...)))))))).....((((((.....)))))).... ( -36.80, z-score = -1.69, R) >droEre2.scaffold_4845 21608477 113 + 22589142 CGUUUCCAGUGGUUCCAUUGGUUCCAGCGUUGUGCAAC-GACUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACAGGCGACUCCUGGUCGCCACCA .(((.((((((....))))))....)))((((((((((-(.........(((((.((((....)))).))..)))......))))))))))).((((((.....)))))).... ( -40.06, z-score = -2.68, R) >droSec1.super_23 121160 104 + 989336 ---------CGCUUGCAGCGGCUCCGGCGUUGUGCAAC-GACUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACAGGCGACUCCUGGUCGCCACCA ---------(((.....))).....((.((((((((((-(.........(((((.((((....)))).))..)))......))))))))))).((((((.....)))))).)). ( -36.26, z-score = -0.90, R) >droSim1.chr2R 18701386 104 + 19596830 ---------CGUUUGCAGCGGCUCCGGCGUUGUGCAAC-GACUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACAGGCGACUCCUGGUCGCCACCA ---------.((((((((((....(((((.((.(((((-(...((((.......))))....)))))))).))))).....))))))).))).((((((.....)))))).... ( -37.10, z-score = -1.27, R) >droMoj3.scaffold_6496 26154041 102 + 26866924 ------UAUUACCAACUGUAUUGUUGCAGUUGUGCAAUUGCCUUUAAAAAGCUAUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACGCAUCGCGGCGCGGCG------ ------......((((((((....)))))))).......(((........((.........))........((((((.((.((((....)))).)).)))))).))).------ ( -27.50, z-score = 0.69, R) >droGri2.scaffold_15245 908864 91 + 18325388 ---------------------UGUUGUUGUUGUGCAAUUGCCUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGAC--GCGACGCGCAGUCGACAGCG ---------------------.(((((((((((((...............((((....)).))(((((..(((.(((........)))))).--))))))))))).))))))). ( -29.50, z-score = -0.38, R) >consensus _________CGCUUGCAGCGGUUCCGGCGUUGUGCAAC_GACUUUAAAAAGCUGUUAACAUGCGUUGUCAUCGCUGCCAUAUGUUGCACGACAGGCGACUCCUGGUCGCCACCA ............................((((((((((...........(((((.((((....)))).))..))).......)))))))))).((((((.....)))))).... (-24.97 = -25.64 + 0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:08 2011