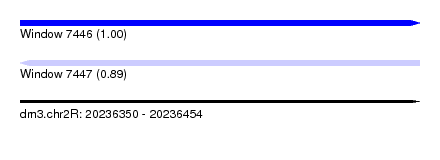
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,236,350 – 20,236,454 |
| Length | 104 |
| Max. P | 0.999456 |

| Location | 20,236,350 – 20,236,454 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.26 |
| Shannon entropy | 0.45033 |
| G+C content | 0.43534 |
| Mean single sequence MFE | -35.36 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.14 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -4.13 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.91 |
| SVM RNA-class probability | 0.999456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
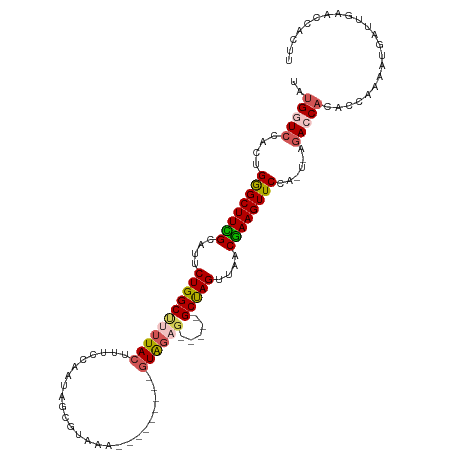
>dm3.chr2R 20236350 104 + 21146708 UAUGAUCCACUGGGCUUCGCCUUCUGGCUUUUACUUGCCACUAGCGUAAA---------GUACAG-----GCUAGUUAACGAAGUUCCA-U-AGACCACACCAAAAUGAAUGGACCACUU ..((.((((.(((((((((....(((((((.(((((..(......)..))---------))).))-----)))))....))))).))))-.-......((......))..)))).))... ( -26.50, z-score = -1.61, R) >droSim1.chr2R 18695550 104 + 19596830 UAUGGUCCACGGAGCUUCGCACUCUGGCUUUUACUUUCCAAUAGCGCUUA---------UUGGAG-----GCUAGUUAACGAAGUUCCA-U-AGACCACACCAAAAUGAUUGAACCACUU ..(((((...(((((((((....((((((.......((((((((...)))---------))))))-----)))))....))))))))).-.-.)))))...................... ( -33.91, z-score = -4.42, R) >droSec1.super_23 115305 104 + 989336 UAUGGUCCACUGGGCUUCGCAAUCUGGCCUUUACUUUCCAAUAGCGUAAA---------GUAGAG-----GCUAGUUAACGAAGUUCCA-U-AGACCACACCAAAAUAAUUGAACCACUU ..(((((...(((((((((....((((((((((((((.(......).)))---------))))))-----)))))....))))).))))-.-.)))))...................... ( -37.40, z-score = -5.66, R) >droYak2.chr2R 20204496 112 + 21139217 UAUGGUCCACCGGGCUUUGUAUUCUGGCGCUUACUUUACAAAAGU-UAAA----UGAUUGUAGAGUAACUGCUAGCUUACAAAGUUCAAAUGGAACCCCUCCAAAAUGACU---CCCUUU ...((((....((((((((((..((((((.(((((((((((....-....----...))))))))))).))))))..))))))))))...((((.....))))....))))---...... ( -38.10, z-score = -5.29, R) >droEre2.scaffold_4845 21602492 118 + 22589142 UGUGGUCCACUGGGCUUUGUAUUCUGGCUCUUACUUUCCAAACGCAUAGAACGGCGAUUGUAGAGUAACUGCCAGCUUGCGAAGUUCCA-UUAGAACACUCCGAAGUGAUUGAACCUUU- ...(((.((.(((((((((((..(((((..(((((((.(((.(((........))).))).)))))))..)))))..))))))).))))-......((((....))))..)).)))...- ( -40.90, z-score = -3.65, R) >consensus UAUGGUCCACUGGGCUUCGCAUUCUGGCUUUUACUUUCCAAUAGCGUAAA_________GUAGAG_____GCUAGUUAACGAAGUUCCA_U_AGACCACACCAAAAUGAUUGAACCACUU ..(((((....((((((((....(((((.....((((........................)))).....)))))....))))))))......)))))...................... (-15.78 = -16.14 + 0.36)

| Location | 20,236,350 – 20,236,454 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.26 |
| Shannon entropy | 0.45033 |
| G+C content | 0.43534 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -13.65 |
| Energy contribution | -13.61 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
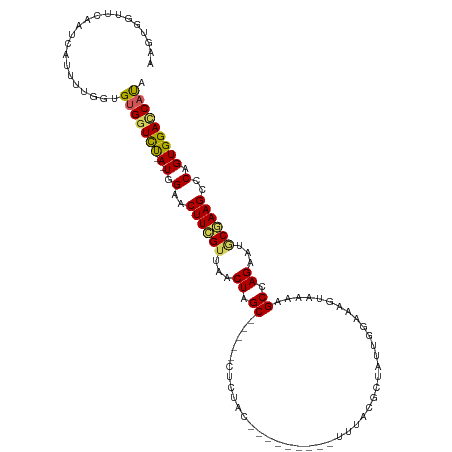
>dm3.chr2R 20236350 104 - 21146708 AAGUGGUCCAUUCAUUUUGGUGUGGUCU-A-UGGAACUUCGUUAACUAGC-----CUGUAC---------UUUACGCUAGUGGCAAGUAAAAGCCAGAAGGCGAAGCCCAGUGGAUCAUA ..(((((((((((((......))))...-.-(((..(((((((.((((((-----.((...---------..)).))))))(((........)))....))))))).)))))))))))). ( -33.80, z-score = -2.23, R) >droSim1.chr2R 18695550 104 - 19596830 AAGUGGUUCAAUCAUUUUGGUGUGGUCU-A-UGGAACUUCGUUAACUAGC-----CUCCAA---------UAAGCGCUAUUGGAAAGUAAAAGCCAGAGUGCGAAGCUCCGUGGACCAUA (((((((...)))))))...((((((((-(-((((.((((((...((.((-----.(((((---------((.....)))))))........)).))...)))))).))))))))))))) ( -36.50, z-score = -3.75, R) >droSec1.super_23 115305 104 - 989336 AAGUGGUUCAAUUAUUUUGGUGUGGUCU-A-UGGAACUUCGUUAACUAGC-----CUCUAC---------UUUACGCUAUUGGAAAGUAAAGGCCAGAUUGCGAAGCCCAGUGGACCAUA .......((((.....))))((((((((-(-(((..((((((...((.((-----((.(((---------(((.((....)).)))))).)))).))...)))))).))).))))))))) ( -34.60, z-score = -2.97, R) >droYak2.chr2R 20204496 112 - 21139217 AAAGGG---AGUCAUUUUGGAGGGGUUCCAUUUGAACUUUGUAAGCUAGCAGUUACUCUACAAUCA----UUUA-ACUUUUGUAAAGUAAGCGCCAGAAUACAAAGCCCGGUGGACCAUA ...(((---..(((...(((((...)))))..))).(((((((..((.((..(((((.(((((...----....-....))))).)))))..)).))..))))))))))((....))... ( -29.90, z-score = -1.51, R) >droEre2.scaffold_4845 21602492 118 - 22589142 -AAAGGUUCAAUCACUUCGGAGUGUUCUAA-UGGAACUUCGCAAGCUGGCAGUUACUCUACAAUCGCCGUUCUAUGCGUUUGGAAAGUAAGAGCCAGAAUACAAAGCCCAGUGGACCACA -...((((((....(((((((((.(((...-.))))))))).)))(((((..(((((...(((.(((........))).)))...)))))..)))))..............))))))... ( -35.30, z-score = -2.10, R) >consensus AAGUGGUUCAAUCAUUUUGGUGUGGUCU_A_UGGAACUUCGUUAACUAGC_____CUCUAC_________UUUACGCUAUUGGAAAGUAAAAGCCAGAAUGCGAAGCCCAGUGGACCAUA .....................(((((((...(((..((((((...((.((..........................................)).))...)))))).)))..))))))). (-13.65 = -13.61 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:07 2011