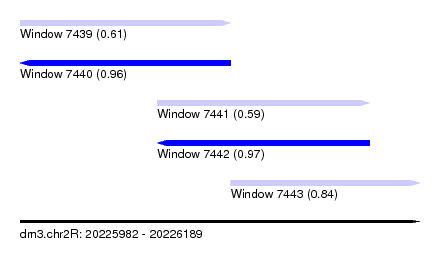
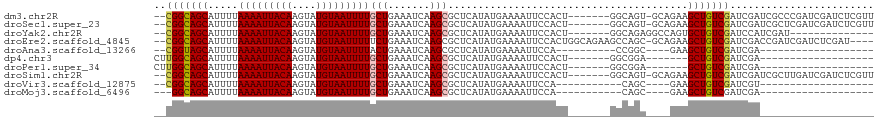
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,225,982 – 20,226,189 |
| Length | 207 |
| Max. P | 0.973128 |

| Location | 20,225,982 – 20,226,091 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.15 |
| Shannon entropy | 0.16369 |
| G+C content | 0.52110 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -28.06 |
| Energy contribution | -27.75 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20225982 109 + 21146708 GGCAAAAGUCAAAGCCGGGCACAAAUUAAAAAUGCCCCACAAUUGCCCGCGAGUUUGAGAUCGGGAACGAGAACGAGAUCGAUCGGGCGAUCGAUCGACAGCUUCUGCA-- .(((.((((.......(((((...........)))))..(.(((((((((((.((((...(((....)))...)))).)))..)))))))).).......)))).))).-- ( -34.80, z-score = -1.53, R) >droSec1.super_23 105060 109 + 989336 GGCAAAAGUCAAAGCCGGGCACAAAUUAAAAAUGCCCCACAAUUGCCCGGGAGCUUGAGAUCGGGAACGGGAACGAGAUCGAUCGAGCGAUCGAUCGACAGCUUCUGCA-- (((((..((....)).(((((...........))))).....)))))((((((((((...(((....)))...)))((((((((....))))))))....)))))))..-- ( -36.80, z-score = -1.95, R) >droEre2.scaffold_4845 21591871 101 + 22589142 GGCAAAAGUCAAAGCCGGGCACAAAUUAAAAAUGCCCCACAAUUGGCCG------CGAGAUCGAGAACGAGAUCGAGAUCGAUC----GGUCGAUCGACAGCUUCUGCGCU (((....(((......(((((...........)))))....((((((((------(((..((((........))))..)))..)----))))))).))).((....))))) ( -32.50, z-score = -1.62, R) >droSim1.chr2R 18684663 109 + 19596830 GGCAAAAGUCAAAGCCGGGCACAAAUUAAAAAUGCCCCACAAUUGCCCGCGAGUUUGAGAUCGGGAACGAGAACGAGAUCGAUCAAGCGAUCGAUCGACAGCUUCUGCA-- .(((.((((((((.(((((((......................)))))).)..)))))..(((....)))......((((((((....)))))))).....))).))).-- ( -32.05, z-score = -1.55, R) >consensus GGCAAAAGUCAAAGCCGGGCACAAAUUAAAAAUGCCCCACAAUUGCCCGCGAGUUUGAGAUCGGGAACGAGAACGAGAUCGAUCGAGCGAUCGAUCGACAGCUUCUGCA__ .(((.((((.......(((((...........))))).................(((...(((....)))...)))((((((((....))))))))....)))).)))... (-28.06 = -27.75 + -0.31)

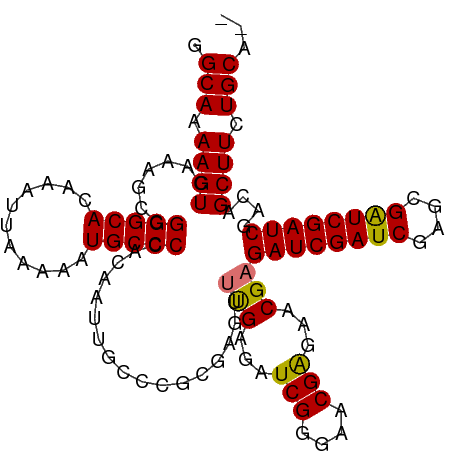
| Location | 20,225,982 – 20,226,091 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.15 |
| Shannon entropy | 0.16369 |
| G+C content | 0.52110 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -33.56 |
| Energy contribution | -33.88 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.960842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20225982 109 - 21146708 --UGCAGAAGCUGUCGAUCGAUCGCCCGAUCGAUCUCGUUCUCGUUCCCGAUCUCAAACUCGCGGGCAAUUGUGGGGCAUUUUUAAUUUGUGCCCGGCUUUGACUUUUGCC --.((((((((.(((((((((((....))))))))........((((.(((........))).)))).......((((((.........)))))))))...).))))))). ( -37.70, z-score = -2.11, R) >droSec1.super_23 105060 109 - 989336 --UGCAGAAGCUGUCGAUCGAUCGCUCGAUCGAUCUCGUUCCCGUUCCCGAUCUCAAGCUCCCGGGCAAUUGUGGGGCAUUUUUAAUUUGUGCCCGGCUUUGACUUUUGCC --.(((((((.....((((((((....))))))))...........((((............))))(((..((.((((((.........)))))).)).))).))))))). ( -37.20, z-score = -1.90, R) >droEre2.scaffold_4845 21591871 101 - 22589142 AGCGCAGAAGCUGUCGAUCGACC----GAUCGAUCUCGAUCUCGUUCUCGAUCUCG------CGGCCAAUUGUGGGGCAUUUUUAAUUUGUGCCCGGCUUUGACUUUUGCC ...(((((((((((.((((((..----(((((....)))))......))))))..)------))))(((..((.((((((.........)))))).)).)))..)))))). ( -37.20, z-score = -2.35, R) >droSim1.chr2R 18684663 109 - 19596830 --UGCAGAAGCUGUCGAUCGAUCGCUUGAUCGAUCUCGUUCUCGUUCCCGAUCUCAAACUCGCGGGCAAUUGUGGGGCAUUUUUAAUUUGUGCCCGGCUUUGACUUUUGCC --.((((((((.(((((((((((....))))))))........((((.(((........))).)))).......((((((.........)))))))))...).))))))). ( -36.80, z-score = -2.15, R) >consensus __UGCAGAAGCUGUCGAUCGAUCGCUCGAUCGAUCUCGUUCUCGUUCCCGAUCUCAAACUCGCGGGCAAUUGUGGGGCAUUUUUAAUUUGUGCCCGGCUUUGACUUUUGCC ...(((((((.....((((((((....))))))))...........((((............))))(((..((.((((((.........)))))).)).))).))))))). (-33.56 = -33.88 + 0.31)

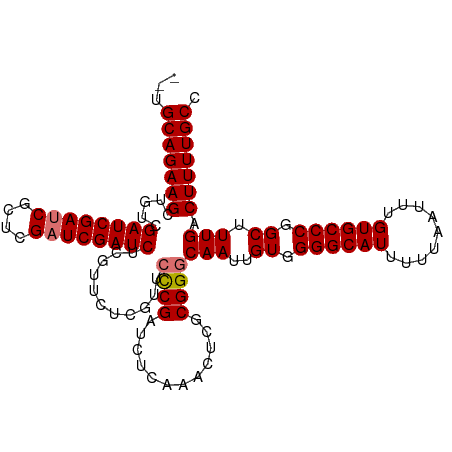
| Location | 20,226,053 – 20,226,163 |
|---|---|
| Length | 110 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Shannon entropy | 0.35870 |
| G+C content | 0.40042 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -14.19 |
| Energy contribution | -14.29 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20226053 110 + 21146708 AACGAGAUCGAUCGGGCGAUCGAUCGACAGCUUCUGC-ACUGCC-------AGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUAAAAUGCUGCCG-- .....((((((((....))))))))(.((((......-...(((-------((((((...))))).)).))(((.(....))))(((((((((....))))))))).....)))).).-- ( -30.10, z-score = -1.66, R) >droSec1.super_23 105131 110 + 989336 AACGAGAUCGAUCGAGCGAUCGAUCGACAGCUUCUGC-ACUGCC-------AGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUAAAAUGCUGCCG-- .....((((((((....))))))))(.((((......-...(((-------((((((...))))).)).))(((.(....))))(((((((((....))))))))).....)))).).-- ( -30.10, z-score = -1.87, R) >droYak2.chr2R 20193908 97 + 21139217 --------------AUCGAUGGAUCGACAGCACUGGCCUCUGCC-------AGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUAAAAUGCUGCCG-- --------------.((((....))))...(((((((....)))-------)))).............((((((.(....))))(((((((((....))))))))).....)))....-- ( -25.90, z-score = -1.98, R) >droEre2.scaffold_4845 21591936 113 + 22589142 ----AUCGAGAUCGAUCGGUCGAUCGACAGCUUCUGC-GCUGGCUUCUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGAAAAAUUACAUACUUGUAAUUUUAAAAUGCUGCCG-- ----.....((((((....))))))(.((((.(((((-((((((....))))))).....(((....)))..........))))(((((((((....))))))))).....)))).).-- ( -34.80, z-score = -2.56, R) >droAna3.scaffold_13266 5699931 85 - 19884421 -------------------UCGAUCGACAGCUUC----GCCGG----------UGGAAUUUUCAUAUGAGCGCUUGAUUUCAGUAAAAUUACAUACUUGUAAUUUUAAAAUGCUACCG-- -------------------..((((((..((((.----....(----------((((...)))))..))))..)))))).((.((((((((((....))))))))))...))......-- ( -16.40, z-score = -0.73, R) >dp4.chr3 1336135 87 - 19779522 -------------------UCGAUCGACAGC-------UCCGCC-------AGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUAAAAUGCUGCCAAG -------------------..((((((..((-------((....-------.(((((...)))))..))))..)))))).(((((((((((((....)))))))).....)))))..... ( -19.40, z-score = -1.54, R) >droPer1.super_34 526245 87 - 916997 -------------------UCGAUCGACAGC-------UCCGCC-------AGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUAAAAUGCUGCCAAG -------------------..((((((..((-------((....-------.(((((...)))))..))))..)))))).(((((((((((((....)))))))).....)))))..... ( -19.40, z-score = -1.54, R) >droSim1.chr2R 18684734 110 + 19596830 AACGAGAUCGAUCAAGCGAUCGAUCGACAGCUUCUGC-ACUGCC-------AGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUAAAAUGCUGCCG-- .....((((((((....))))))))(.((((......-...(((-------((((((...))))).)).))(((.(....))))(((((((((....))))))))).....)))).).-- ( -29.60, z-score = -2.01, R) >droVir3.scaffold_12875 7337733 84 + 20611582 -------------------ACGAUCGACAGCUUC----GCUG-----------UGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUAAAAUGCUGCCG-- -------------------..((((((..(((.(----(.((-----------((((...)))))))))))..)))))).(((((((((((((....)))))))).....)))))...-- ( -20.00, z-score = -1.73, R) >droMoj3.scaffold_6496 26132384 83 + 26866924 -------------------UCGAUCGACAGCUUC----GCUG-----------UGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUAAAAUGCUGCC--- -------------------..((((((..(((.(----(.((-----------((((...)))))))))))..)))))).(((((((((((((....)))))))).....)))))..--- ( -20.00, z-score = -1.81, R) >consensus ___________________UCGAUCGACAGCUUC____GCUGCC_______AGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUUGUAAUUUUAAAAUGCUGCCG__ .........................(.((((......................((((((((((....))).....)))))))..(((((((((....))))))))).....)))).)... (-14.19 = -14.29 + 0.10)
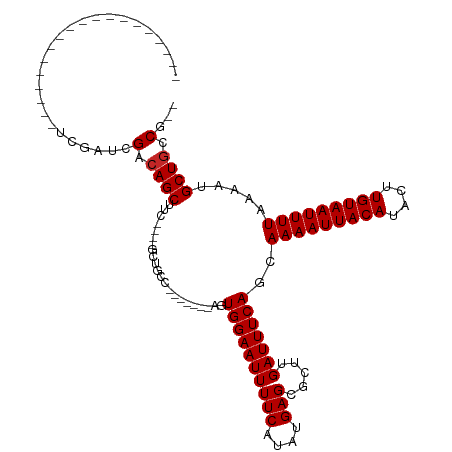
| Location | 20,226,053 – 20,226,163 |
|---|---|
| Length | 110 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.08 |
| Shannon entropy | 0.35870 |
| G+C content | 0.40042 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.973128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20226053 110 - 21146708 --CGGCAGCAUUUUAAAAUUACAAGUAUGUAAUUUUGCUGAAAUCAAGCGCUCAUAUGAAAAUUCCACU-------GGCAGU-GCAGAAGCUGUCGAUCGAUCGCCCGAUCGAUCUCGUU --.((((((....((((((((((....))))))))))..........((((((...((.......))..-------.).)))-))....))))))((((((((....))))))))..... ( -35.20, z-score = -3.28, R) >droSec1.super_23 105131 110 - 989336 --CGGCAGCAUUUUAAAAUUACAAGUAUGUAAUUUUGCUGAAAUCAAGCGCUCAUAUGAAAAUUCCACU-------GGCAGU-GCAGAAGCUGUCGAUCGAUCGCUCGAUCGAUCUCGUU --.((((((....((((((((((....))))))))))..........((((((...((.......))..-------.).)))-))....))))))((((((((....))))))))..... ( -35.20, z-score = -3.09, R) >droYak2.chr2R 20193908 97 - 21139217 --CGGCAGCAUUUUAAAAUUACAAGUAUGUAAUUUUGCUGAAAUCAAGCGCUCAUAUGAAAAUUCCACU-------GGCAGAGGCCAGUGCUGUCGAUCCAUCGAU-------------- --(((((((((((((((((((((....)))))))))(((.......))).........)))))...(((-------(((....)))))))))))))..........-------------- ( -29.40, z-score = -2.95, R) >droEre2.scaffold_4845 21591936 113 - 22589142 --CGGCAGCAUUUUAAAAUUACAAGUAUGUAAUUUUUCUGAAAUCAAGCGCUCAUAUGAAAAUUCCACUGGCAGAAGCCAGC-GCAGAAGCUGUCGAUCGACCGAUCGAUCUCGAU---- --.((((((.(((.(((((((((....)))))))))...))).....((((((....)).........((((....))))))-))....))))))((((((....)))))).....---- ( -34.70, z-score = -3.18, R) >droAna3.scaffold_13266 5699931 85 + 19884421 --CGGUAGCAUUUUAAAAUUACAAGUAUGUAAUUUUACUGAAAUCAAGCGCUCAUAUGAAAAUUCCA----------CCGGC----GAAGCUGUCGAUCGA------------------- --((..(((....((((((((((....))))))))))...........((((...((....))....----------..)))----)..)))..)).....------------------- ( -18.00, z-score = -1.19, R) >dp4.chr3 1336135 87 + 19779522 CUUGGCAGCAUUUUAAAAUUACAAGUAUGUAAUUUUGCUGAAAUCAAGCGCUCAUAUGAAAAUUCCACU-------GGCGGA-------GCUGUCGAUCGA------------------- .((((((((((((((((((((((....)))))))))(((.......))).........)))))(((...-------...)))-------))))))))....------------------- ( -21.90, z-score = -1.47, R) >droPer1.super_34 526245 87 + 916997 CUUGGCAGCAUUUUAAAAUUACAAGUAUGUAAUUUUGCUGAAAUCAAGCGCUCAUAUGAAAAUUCCACU-------GGCGGA-------GCUGUCGAUCGA------------------- .((((((((((((((((((((((....)))))))))(((.......))).........)))))(((...-------...)))-------))))))))....------------------- ( -21.90, z-score = -1.47, R) >droSim1.chr2R 18684734 110 - 19596830 --CGGCAGCAUUUUAAAAUUACAAGUAUGUAAUUUUGCUGAAAUCAAGCGCUCAUAUGAAAAUUCCACU-------GGCAGU-GCAGAAGCUGUCGAUCGAUCGCUUGAUCGAUCUCGUU --.((((((....((((((((((....))))))))))..........((((((...((.......))..-------.).)))-))....))))))((((((((....))))))))..... ( -34.30, z-score = -2.85, R) >droVir3.scaffold_12875 7337733 84 - 20611582 --CGGCAGCAUUUUAAAAUUACAAGUAUGUAAUUUUGCUGAAAUCAAGCGCUCAUAUGAAAAUUCCA-----------CAGC----GAAGCUGUCGAUCGU------------------- --(((((((....((((((((((....))))))))))...........((((...((....))....-----------.)))----)..))))))).....------------------- ( -22.20, z-score = -2.41, R) >droMoj3.scaffold_6496 26132384 83 - 26866924 ---GGCAGCAUUUUAAAAUUACAAGUAUGUAAUUUUGCUGAAAUCAAGCGCUCAUAUGAAAAUUCCA-----------CAGC----GAAGCUGUCGAUCGA------------------- ---((((((....((((((((((....))))))))))...........((((...((....))....-----------.)))----)..))))))......------------------- ( -20.40, z-score = -1.79, R) >consensus __CGGCAGCAUUUUAAAAUUACAAGUAUGUAAUUUUGCUGAAAUCAAGCGCUCAUAUGAAAAUUCCACU_______GGCAGC____GAAGCUGUCGAUCGA___________________ ..(((((((.....(((((((((....)))))))))(((.......)))........................................)))))))........................ (-17.98 = -18.05 + 0.07)
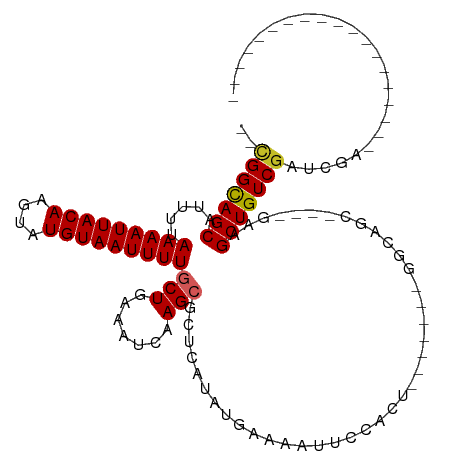
| Location | 20,226,091 – 20,226,189 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.24 |
| Shannon entropy | 0.36773 |
| G+C content | 0.38026 |
| Mean single sequence MFE | -24.89 |
| Consensus MFE | -14.89 |
| Energy contribution | -15.96 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20226091 98 + 21146708 -----CUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUU-GUAAUUUUAAAAUGCUGCC---GAGUGCCGAGAGAGUUGAAAAGCAACA----- -----.(((...(..(((((((...((.(((((((....(((((((((((((....)-))))))).....))))).)---)))))))).)))))))..)..)))...----- ( -26.00, z-score = -2.16, R) >droSec1.super_23 105169 98 + 989336 -----CUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUU-GUAAUUUUAAAAUGCUGCC---GAGUGCCGAGAGAGUUGAAAAGCAACA----- -----.(((...(..(((((((...((.(((((((....(((((((((((((....)-))))))).....))))).)---)))))))).)))))))..)..)))...----- ( -26.00, z-score = -2.16, R) >droYak2.chr2R 20193928 103 + 21139217 GGCCUCUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUU-GUAAUUUUAAAAUGCUGCC---GAGUGCCGAGAGAGUUGAGCAGCAGCA----- .....((((..((..(((((((...((.(((((((....(((((((((((((....)-))))))).....))))).)---)))))))).)))))))..)).))))..----- ( -30.50, z-score = -1.54, R) >droEre2.scaffold_4845 21591972 103 + 22589142 GGCUUCUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGAAAAAUUACAUACUU-GUAAUUUUAAAAUGCUGCC---GAGUGCCGAGAGAGUUGAAAAGCAACA----- (((....))).((....((((((.....(((((((.(..((..(((((((((....)-))))))))....))..).)---))))))(....)...))))))...)).----- ( -25.40, z-score = -1.24, R) >droAna3.scaffold_13266 5699944 97 - 19884421 -----UCGCCGGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGUAAAAUUACAUACUU-GUAAUUUUAAAAUGCUACC---GAGUGCCGAGAGAGUUGAA-AGCAACA----- -----..((...(..(((((((...((.(((((((.(..((.((((((((((....)-)))))))))...))..).)---)))))))).)))))))..)-.))....----- ( -23.90, z-score = -1.87, R) >dp4.chr3 1336148 99 - 19779522 -----CCGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUU-GUAAUUUUAAAAUGCUGCCAA-GAGUGCCAAGAGAGUUGAA-ACCAACA----- -----(((....)))(((((((...((.((((((.....(((((((((((((....)-))))))).....)))))....-)))))))).)))))))...-.......----- ( -25.50, z-score = -2.83, R) >droPer1.super_34 526258 99 - 916997 -----CCGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUU-GUAAUUUUAAAAUGCUGCCAA-GAGUGCCAAGAGAGUUGAA-ACCAACA----- -----(((....)))(((((((...((.((((((.....(((((((((((((....)-))))))).....)))))....-)))))))).)))))))...-.......----- ( -25.50, z-score = -2.83, R) >droSim1.chr2R 18684772 98 + 19596830 -----CUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUU-GUAAUUUUAAAAUGCUGCC---GAGUGCCGAGAGAGUUGAAAAGCAACA----- -----.(((...(..(((((((...((.(((((((....(((((((((((((....)-))))))).....))))).)---)))))))).)))))))..)..)))...----- ( -26.00, z-score = -2.16, R) >droWil1.scaffold_180745 1289097 98 + 2843958 -----CAGUCUGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGUGAAAUUACAUACUUUGUAAUUUUAAAAUGCUACC---AACUGCCGAGAGAGUUGAA-GCUCACA----- -----.....((((((...))))))((((((((((....)))((((((((((.....))))))))))....))...(---((((..(....))))))..-)))))..----- ( -21.50, z-score = -0.94, R) >droVir3.scaffold_12875 7337746 92 + 20611582 ------UCGCUGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUU-GUAAUUUUAAAAUGCUGCC---GAGUGCC--GAGUGCUGAGUUGCA-------- ------......((.(((((.(((.((.(((((((....(((((((((((((....)-))))))).....))))).)---)))))))--).)))..))))).))-------- ( -26.00, z-score = -1.66, R) >droMoj3.scaffold_6496 26132397 78 + 26866924 ------UCGCUGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUU-GUAAUUUUAAAAUGCUGCC---GAGUUGCA------------------------ ------(((.((((((...)))))))))(((((((....(((((((((((((....)-))))))).....))))).)---))).))).------------------------ ( -19.30, z-score = -1.54, R) >droGri2.scaffold_15245 884712 101 + 18325388 ----------GCUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUU-GUAAUUUUAAAAUGCUGACUUCCAGCAACAAGGAAUUAAAUCAAAUAUAUAUCU ----------(((((((..(((....))).........((((((((((((((....)-))))))).....)))))).)))))))............................ ( -23.10, z-score = -2.87, R) >consensus _____CUGCCAGUGGAAUUUUCAUAUGAGCGCUUGAUUUCAGCAAAAUUACAUACUU_GUAAUUUUAAAAUGCUGCC___GAGUGCCGAGAGAGUUGAA_AGCAACA_____ ...............(((((((...((.((((((.....((((((((((((.......))))))).....))))).....)))))))).)))))))................ (-14.89 = -15.96 + 1.07)
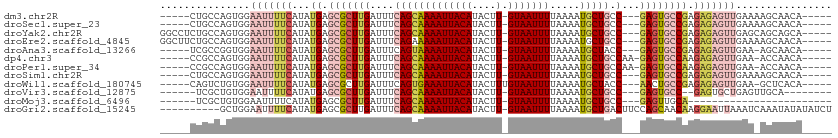

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:04 2011