
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,225,187 – 20,225,295 |
| Length | 108 |
| Max. P | 0.998849 |

| Location | 20,225,187 – 20,225,295 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 56.64 |
| Shannon entropy | 0.88613 |
| G+C content | 0.44995 |
| Mean single sequence MFE | -32.64 |
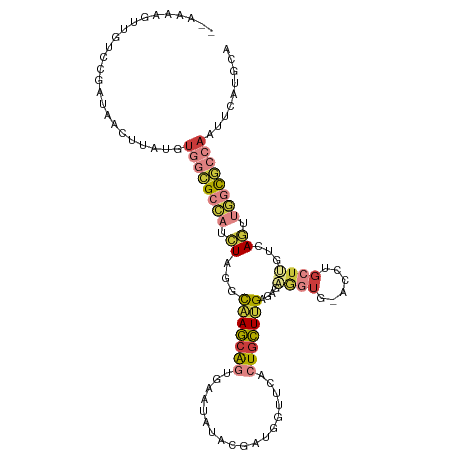
| Consensus MFE | -14.30 |
| Energy contribution | -15.76 |
| Covariance contribution | 1.46 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965676 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
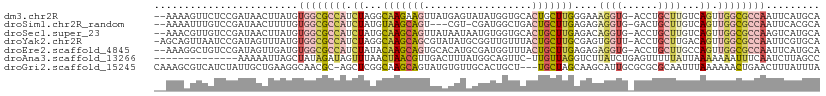
>dm3.chr2R 20225187 108 + 21146708 --AAAAGUUCUCCGAUAACUUAUGUGGCGCCAUCUAGGCAAGAAGUUAUGAGUAUAUGGUGCACUGCUUGGGAAAGGUG-ACCUGCUUGUCAGUUGGCGCCAAUUCAUGCA --..(((((.......)))))...((((((((.((.((((((..(((((..((((...))))....(((....))))))-))...)))))))).))))))))......... ( -31.30, z-score = -0.44, R) >droSim1.chr2R_random 2989290 104 + 2996586 --AAAAUUUGUCCGAUAACUUUUGUGGCGCCAUCUAUGUAAGCAGU---CGU-CGAUGGCUGACUGCUUGAGAGAGGUG-GACUGCUUGUCAGUUGGCGCCAAUUCACGCA --......................((((((((....((((((((((---(((-((.....))))..(((....)))...-))))))))).))..))))))))......... ( -37.00, z-score = -2.14, R) >droSec1.super_23 104283 108 + 989336 --AAACGUUGUCCGAUAACUUAUGUGGCGCCAUCUAUGCAAGCAGUUAUAAUAAUGUGGUGCACUGCUUGAGACAGGUG-ACCUGCUUGUCAGUUGGCGCCAAGUCAUGCA --....(((((...))))).((((((((((((.((...(((((((((((.........))).)))))))).(((((((.-....))))))))).))))))))...)))).. ( -37.70, z-score = -1.91, R) >droYak2.chr2R 20193026 109 + 21139217 -AGCAGUUAAUCCGAUAGUUUAUGUGGCGCCAUCUAGGCAAGCAGCGUAUAUGCGGUUGUUUACUGCUUGCGAGUGGUU-ACCUGCUUGACAGUUGGCGCCAAUUCGUGCA -.(((.......(((.........((((((((.((...(((((((((((...(((((.....))))).)))).......-..)))))))..)).))))))))..)))))). ( -37.21, z-score = -0.80, R) >droEre2.scaffold_4845 21591159 108 + 22589142 --AAAGGCUGUCCGAUAGUUGAUGUGGCGCCAUCUAUACAAGCAGUGCACAUGCGAUGGUUUACUGCUUGAGAGAGGUG-ACCUGCUUGCCAGUUGGCGCCAAUUCAUGCA --...((((((...))))))((..(((((((((((...(((((((((.((........)).)))))))))..)))((..-(.....)..))...))))))))..))..... ( -38.50, z-score = -1.24, R) >droAna3.scaffold_13266 5698876 96 - 19884421 --------------AAAAAUUAGCUAUAGAUAGUUUAACUAACGUUGACUUUAUGGCAGUUC-UUGUUAGGUCUUAUCUGAGUUUUUAUUAAAAAAAUUUCAAUCUUAGCC --------------........(((((((((((.....((((((..((((.......)))).-.))))))...)))))))(((((((.....))))))).......)))). ( -16.50, z-score = -0.83, R) >droGri2.scaffold_15245 883698 107 + 18325388 CAAAGCGUCAUCUAUUGCUGAAGGCAACGC-AGCUCGGCAAGCAGUAUGUGUUGCACUGCU---UGCUAGCAAGCAUUGCGCGCGCAAUUUAAAAAACUGAACUUUAUUUA ....(((((((.(((((((...(....)((-......)).))))))).)))..(((.((((---((....)))))).)))))))........................... ( -30.30, z-score = 0.47, R) >consensus __AAAAGUUGUCCGAUAACUUAUGUGGCGCCAUCUAGGCAAGCAGUGAAUAUACGAUGGUUCACUGCUUGAGAGAGGUG_ACCUGCUUGUCAGUUGGCGCCAAUUCAUGCA ........................((((((((.((...(((((((..................)))))))....((((......))))...)).))))))))......... (-14.30 = -15.76 + 1.46)
| Location | 20,225,187 – 20,225,295 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 56.64 |
| Shannon entropy | 0.88613 |
| G+C content | 0.44995 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -15.32 |
| Energy contribution | -16.25 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.998849 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
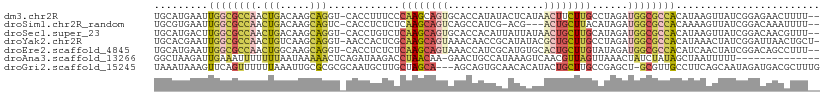
>dm3.chr2R 20225187 108 - 21146708 UGCAUGAAUUGGCGCCAACUGACAAGCAGGU-CACCUUUCCCAAGCAGUGCACCAUAUACUCAUAACUUCUUGCCUAGAUGGCGCCACAUAAGUUAUCGGAGAACUUUU-- .........((((((((.(((.((((..(((-((((((....)))..))).))).(((....)))....))))..))).))))))))...(((((.......)))))..-- ( -24.00, z-score = -0.25, R) >droSim1.chr2R_random 2989290 104 - 2996586 UGCGUGAAUUGGCGCCAACUGACAAGCAGUC-CACCUCUCUCAAGCAGUCAGCCAUCG-ACG---ACUGCUUACAUAGAUGGCGCCACAAAAGUUAUCGGACAAAUUUU-- .........((((((((.(((..((((((((-...((......))..(((.......)-)))---)))))))...))).)))))))).....(((....))).......-- ( -30.30, z-score = -2.17, R) >droSec1.super_23 104283 108 - 989336 UGCAUGACUUGGCGCCAACUGACAAGCAGGU-CACCUGUCUCAAGCAGUGCACCACAUUAUUAUAACUGCUUGCAUAGAUGGCGCCACAUAAGUUAUCGGACAACGUUU-- .(.((((((((((((((.(((....(((((.-..)))))..((((((((................))))))))..))).))))))))....)))))))...........-- ( -35.39, z-score = -2.72, R) >droYak2.chr2R 20193026 109 - 21139217 UGCACGAAUUGGCGCCAACUGUCAAGCAGGU-AACCACUCGCAAGCAGUAAACAACCGCAUAUACGCUGCUUGCCUAGAUGGCGCCACAUAAACUAUCGGAUUAACUGCU- .(((.....((((((((.(((.(((((((((-....((((....).))).....)))((......))))))))..))).))))))))...........(......)))).- ( -32.90, z-score = -1.94, R) >droEre2.scaffold_4845 21591159 108 - 22589142 UGCAUGAAUUGGCGCCAACUGGCAAGCAGGU-CACCUCUCUCAAGCAGUAAACCAUCGCAUGUGCACUGCUUGUAUAGAUGGCGCCACAUCAACUAUCGGACAGCCUUU-- ....(((..((((((((.(((((((((((((-...((((....)).))...)))...((....))..))))))).))).))))))))..)))......((....))...-- ( -33.80, z-score = -1.27, R) >droAna3.scaffold_13266 5698876 96 + 19884421 GGCUAAGAUUGAAAUUUUUUUAAUAAAAACUCAGAUAAGACCUAACAA-GAACUGCCAUAAAGUCAACGUUAGUUAAACUAUCUAUAGCUAAUUUUU-------------- (((((..((((((.....))))))........(((((....(((((..-..(((.......)))....)))))......))))).))))).......-------------- ( -12.10, z-score = -0.30, R) >droGri2.scaffold_15245 883698 107 - 18325388 UAAAUAAAGUUCAGUUUUUUAAAUUGCGCGCGCAAUGCUUGCUAGCA---AGCAGUGCAACACAUACUGCUUGCCGAGCU-GCGUUGCCUUCAGCAAUAGAUGACGCUUUG ....((((((...(((.((((..((((....(((((((..(((.(((---(((((((.......))))))))))..))).-))))))).....)))))))).))))))))) ( -37.30, z-score = -2.26, R) >consensus UGCAUGAAUUGGCGCCAACUGACAAGCAGGU_CACCUCUCUCAAGCAGUGAACCAUCGAAUAAUCACUGCUUGCAUAGAUGGCGCCACAUAAAUUAUCGGACAACCUUU__ .........((((((((.(((.....)))............((((((((................))))))))......))))))))........................ (-15.32 = -16.25 + 0.93)
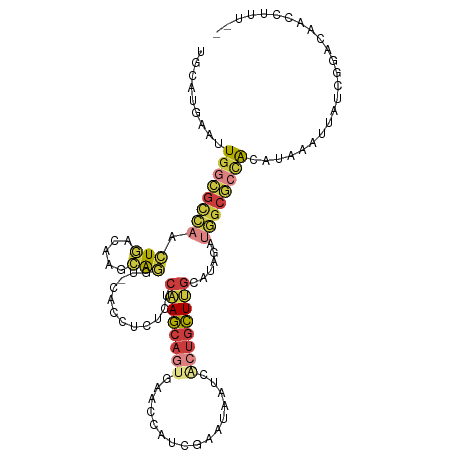
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:48:00 2011