| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,214,728 – 20,214,824 |
| Length | 96 |
| Max. P | 0.728362 |

| Location | 20,214,728 – 20,214,824 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 71.29 |
| Shannon entropy | 0.53987 |
| G+C content | 0.45037 |
| Mean single sequence MFE | -25.31 |
| Consensus MFE | -12.83 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20214728 96 - 21146708 AGGAAUAUGUUUGUGCAUUU-GUGGCGGUGGAAUGUGUAUGUCUGGAUAUGUGUAUCUGC---GAUC--UGACGCUGUUU----AGACGGUGUCUAUUUCCCAUUC-- .((((.((((....))))..-((.((((....(((..(((((....)))))..)))))))---.)).--.((((((((..----..))))))))...)))).....-- ( -23.20, z-score = -0.29, R) >droSim1.chr2R 18662647 96 - 19596830 AGGGAAAUGUUUGUGCAUUU-GUGGCGGUGGAAUGUGUAUGUCUGGAUAUGUGUAUCUGC---GAUC--UGACGCUGUUU----AGACGGUGUCUAUUUCCCCUUC-- .((((((((....(((....-...)))(..((.((..(((((....)))))..))))..)---....--.((((((((..----..))))))))))))))))....-- ( -29.20, z-score = -2.49, R) >droSec1.super_23 93867 96 - 989336 AGGGAAAUGUUUGUGCAUUU-GUGGCGGUGGAAUGUGUAUGUCUGGAUAUGUGUAUCUGC---GAUC--CGACGCUGUUU----AUACGGUGUCUAUUUCCCCUUC-- .((((((((...........-..((..(..((.((..(((((....)))))..))))..)---...)--)((((((((..----..))))))))))))))))....-- ( -30.40, z-score = -2.66, R) >droYak2.chr2R 20181954 92 - 21139217 AGGGAAAUGUUUGUGCAUUU-GCGGCAGUGGA----AUGUGUCUGGAUGUGUGUAUCUGC---GAUC--UGACGCUGUUU----AGACGGUGUCUAUUUCUCCUUC-- .((((((((....((((..(-((((((.((((----.....))))..))).))))..)))---)...--.((((((((..----..))))))))))))))))....-- ( -25.60, z-score = -1.32, R) >droEre2.scaffold_4845 21580736 92 - 22589142 AGGGAAACGUUUGUGCAUUU-GUGGCAGUGGA----AUGUGUCUGGAUGUGUGUAUCUGC---GAUC--UGGCGCUGUUU----AGACGGUGUCUAUUUCUCCUUC-- .(((((((((..((((((..-..((((.....----...)))).......))))))..))---)...--.((((((((..----..))))))))..))))))....-- ( -24.80, z-score = -0.98, R) >droAna3.scaffold_13266 5688362 85 + 19884421 AAAGAAAUGUUUGUGCAUUG-CGAGCGGAAUG-UCUGGAUAUCUGGGUA------UCUGC---GAUCUCUGACUCUGUUU----AGACGGUGUAUGUUUC-------- ...(((((....((((((((-...(((((.(.-.(..(....)..)..)------)))))---....(((((......))----))))))))))))))))-------- ( -20.90, z-score = -0.98, R) >dp4.chr3 1320863 86 + 19779522 AGAGAAAUGUUUGUGCAUUG-CCAAUGGAGGA----AUGUGUCUGGAUA-------CUGC---GAUCUGUGGCACUGUUU----AGACGGUGUCUACUUCUCCAU--- .....(((((....))))).-...((((((((----..((((....)))-------)...---.....((((((((((..----..)))))).))))))))))))--- ( -24.10, z-score = -1.00, R) >droPer1.super_34 509902 86 + 916997 AGAGAAAUGUUUGUGCAUUG-CCAAUGGAGGA----AUGUGUCUGGAUA-------CUGC---GAUCUGUGGCACUGUUU----AGACGGUGUCUACUUCUCCAU--- .....(((((....))))).-...((((((((----..((((....)))-------)...---.....((((((((((..----..)))))).))))))))))))--- ( -24.10, z-score = -1.00, R) >droWil1.scaffold_180745 1250153 107 - 2843958 AAGAAAAUGUUGGUGCAUUGUUUGACAAGUGGGUGAAGGGAUAGGGGCA-AUGUGUUUGCUUUGUUUUUUUCUUCUGUCUCUGGAUGCGGUGUCUACUUUUCUUUCCU (((((((.((.((.(((((((......((.((..((((..(..((((((-(.....))))))).....)..))))..)).))....)))))))))))))))))).... ( -25.50, z-score = -0.72, R) >consensus AGGGAAAUGUUUGUGCAUUU_GUGGCGGUGGA_U__AUGUGUCUGGAUAUGUGUAUCUGC___GAUC__UGACGCUGUUU____AGACGGUGUCUAUUUCUCCUUC__ .(((((((((....)))))...................................................(((((((((......)))))))))....))))...... (-12.83 = -13.27 + 0.44)

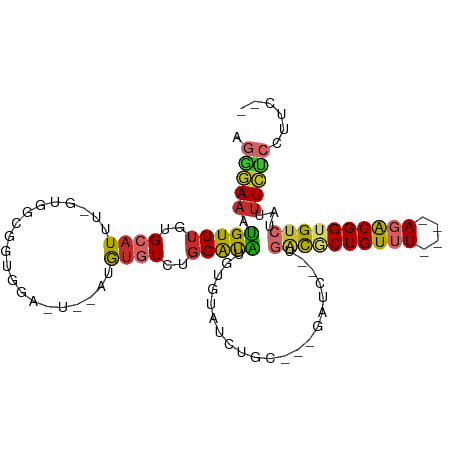

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:59 2011