| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,192,693 – 20,192,804 |
| Length | 111 |
| Max. P | 0.514071 |

| Location | 20,192,693 – 20,192,804 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.24 |
| Shannon entropy | 0.25593 |
| G+C content | 0.44106 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -18.42 |
| Energy contribution | -18.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
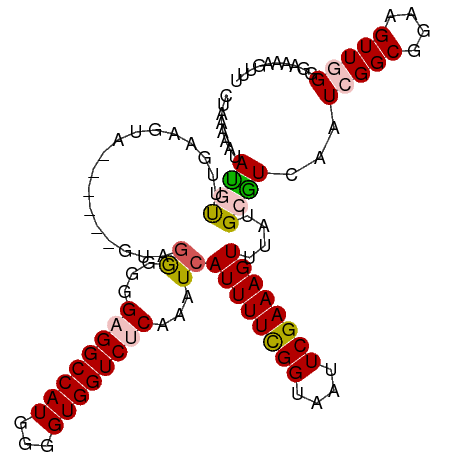
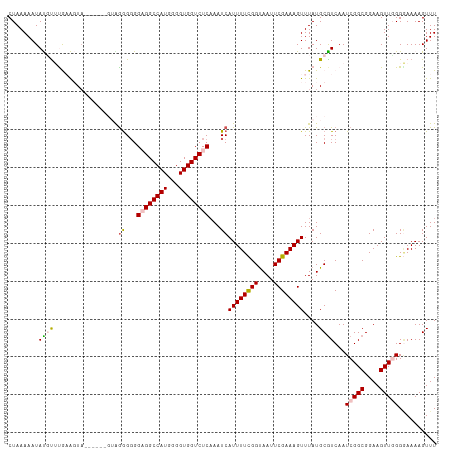
>dm3.chr2R 20192693 111 - 21146708 CUAAAAAUAUGUUCAAAGUA--GGGGGGGGGGGGGAGGCCAUGGGGUGGUCUCAAAUCAUUUUCGGCAAUUCGAAAGUUUAUGCAUCAAUCGGCGGAAGUUGGGGAAAAGUUU ...........(((......--(..((..((...((((((((...))))))))...)).))..).(((...(....)....))).....(((((....))))).)))...... ( -25.50, z-score = -1.65, R) >droSim1.chr2R 18654217 106 - 19596830 CUAAAAAUAUGUUCGAAGUA-------UAGGGGGGAGGCCAUGGGGUGGUCUCAAAUCAUUUUCGGUAAUUCGAAAGUUUAUGCGUCAAUCGGCGGAAGUUCGGGAAAAGUUU ...........(((((.(((-------(((((..((((((((...))))))))...))((((((((....)))))))))))))).))..(((((....).)))))))...... ( -27.90, z-score = -2.70, R) >droSec1.super_23 72058 106 - 989336 CUAAAAAUAUGUUUGAAGUA-------UAGGGGGGAGGCCAUGGGGUGGUCUCAAAUCAUUUUCGGUAAUUCGAAAGUUUAUGCGUCAAUCGGCGGAAGUUCGGGAAAAGUUU ............((((.(((-------(((((..((((((((...))))))))...))((((((((....)))))))))))))).))))(((((....).))))......... ( -28.30, z-score = -3.05, R) >droYak2.chr2R 20158872 113 - 21139217 CUAAAAAUACUCGUAUGUUCCCAGAGGAGGGGUGGUGGCCAUGGGGUGGUCUCCAAUCAUUUUUGGUAAUUCGAAAGUUUAUGCGUCAAUCGGCGGAAGUUGGGCAAAAGUUU ....((((..(((...(((.(((((((.((..(((.((((((...)))))).))).)).))))))).))).)))..)))).(((.....(((((....))))))))....... ( -29.40, z-score = -0.55, R) >droEre2.scaffold_4845 21558332 110 - 22589142 CUAAAAAUACCCGUGUGUUA---AAAGUAAAGGGGUGGCCAUGGGGUGGUCUCAAAUCAUUUUCGGUAAUUCGAAAGUUUAUGCGUCAAUCGGCGGAAGUUGGGAAAAAGUUU .........((((((.((((---............))))))))))....((((((.((((((((((....)))))))).....((((....))))))..))))))........ ( -25.60, z-score = -1.21, R) >consensus CUAAAAAUAUGUUUGAAGUA______GUAGGGGGGAGGCCAUGGGGUGGUCUCAAAUCAUUUUCGGUAAUUCGAAAGUUUAUGCGUCAAUCGGCGGAAGUUGGGGAAAAGUUU ........((((.................((...((((((((...))))))))...))((((((((....))))))))....))))...(((((....))))).......... (-18.42 = -18.86 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:57 2011