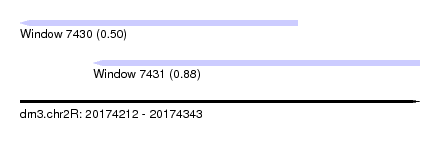
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,174,212 – 20,174,343 |
| Length | 131 |
| Max. P | 0.880916 |

| Location | 20,174,212 – 20,174,303 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 94.14 |
| Shannon entropy | 0.08073 |
| G+C content | 0.61418 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2R 20174212 91 - 21146708 AGGCACGCCACCGCCUUGCCAAUCGAAGGGCCACACGACCACCCACAGCCACAUCCACAUCCACUGCCAUCCGAAUGGGAGAUGGCAGACA .(((........(((((.........))))).....(......)...))).............(((((((((......).))))))))... ( -23.30, z-score = -0.51, R) >droSim1.chr2R 18635666 85 - 19596830 AGGCACGCCACCGCCUUGCCAAUCGAAGGGCCACACGACUACCCACAGCCAC------UUCCACUGCCAUCCGAAUGGGAGAUGGCAGACA .((((.((....))..))))....(((((((................))).)------)))..(((((((((......).))))))))... ( -26.89, z-score = -2.04, R) >droSec1.super_23 53639 91 - 989336 AGGCACGCCACCGCCUUGCCAAUCGAAGGGCCACACGACCACCCACAGCCACAUCCACAUCCACUGCCAUCCGAAUGGGAGAUGGCAGACA .(((........(((((.........))))).....(......)...))).............(((((((((......).))))))))... ( -23.30, z-score = -0.51, R) >consensus AGGCACGCCACCGCCUUGCCAAUCGAAGGGCCACACGACCACCCACAGCCACAUCCACAUCCACUGCCAUCCGAAUGGGAGAUGGCAGACA .(((........(((((.........))))).....(......)...))).............(((((((((......).))))))))... (-23.30 = -23.30 + 0.00)


| Location | 20,174,236 – 20,174,343 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.39 |
| Shannon entropy | 0.35713 |
| G+C content | 0.57794 |
| Mean single sequence MFE | -31.41 |
| Consensus MFE | -22.39 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.880916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20174236 107 - 21146708 GUGGAUACAGCUGCAACUGGCUGUGUUUUCACAGCCAUUCAGGCACGCCACCGCCUUGCCAAUCGAAGGGCCACACGACCACCCACAGCCACAUCCACAUCCACUGC-- ((((((...((((....((((((((....))))))))....((((.((....))..)))).......(((...........))).))))...)))))).........-- ( -36.40, z-score = -3.12, R) >droSim1.chr2R 18635690 101 - 19596830 GUGGAUACAGCUGCAACUGGCUGUGUUUUCACAUCCAUUCAGGCACGCCACCGCCUUGCCAAUCGAAGGGCCACACGACUACCCACAGCCACUUCCACUGC-------- (((((....((((....((((((((....)))).......((((........)))).))))......(((...........))).))))....)))))...-------- ( -28.70, z-score = -1.26, R) >droSec1.super_23 53663 107 - 989336 GUGGAUACAGCUGCAACUGGCUGUGUUUUCACAGCCAUUCAGGCACGCCACCGCCUUGCCAAUCGAAGGGCCACACGACCACCCACAGCCACAUCCACAUCCACUGC-- ((((((...((((....((((((((....))))))))....((((.((....))..)))).......(((...........))).))))...)))))).........-- ( -36.40, z-score = -3.12, R) >droYak2.chr2R 20140197 101 - 21139217 GUGGAUACAGCUGCAACUGGCUGUGUUUUCACAGCCAUUCAGGCACGCCACCGCCUUGCCAAUCGAAGGGCCACACGACCACCCAGAGCCACCUCCACUGC-------- (((((....((((((..((((((((....))))))))...((((........)))))))........(((...........)))..)))....)))))...-------- ( -32.90, z-score = -1.63, R) >droEre2.scaffold_4845 21538488 101 - 22589142 GUGGAUACAGCUGCAACUGGCUGUGUUUUCACAGCCAUUCAGGCACGCCACCGCCUUGCCAAUCGAAGGGCCACACGACCACCCAGAGCCACCUCCACUGC-------- (((((....((((((..((((((((....))))))))...((((........)))))))........(((...........)))..)))....)))))...-------- ( -32.90, z-score = -1.63, R) >droAna3.scaffold_13266 5646924 96 + 19884421 GUGGAUACAGCUGCAACUGGCUGUGUUUUCACAGCCAUUCAGGCAAGCCACCGCCAUGCCAAUCGAAGGAACACACGCUCCUCCCGCCCCAUCCUC------------- ((((....(((.(((..((((((((....))))))))....(((........))).)))...((....))......)))....)))).........------------- ( -29.10, z-score = -2.04, R) >dp4.chr3 1279379 109 + 19779522 GUGGAUACAGCUGCAACUGGGUGUGUUUUCACAGCCAUUCAGGCACGCCACCGCCAUGCCAAUCGAAGGGACACACAGAUACAGUGAGGCACACAGCGAGACAGACAGA ((((.....(((.....(((.((((....)))).)))....)))...))))(((..((((..((....))...(((.......))).))))....)))........... ( -28.90, z-score = 0.05, R) >droWil1.scaffold_180745 1149686 100 - 2843958 GUGGAUACAGCUGCAACUGGCUGUGUUUUCACAGCCAUUCAGGCACGCCACCGCCAUGCCAAUCGAAGGGACACACACACAUAG------ACACAGCCAAACAUAC--- (..(......)..)...(((((((((((.............((((.((....))..))))..((....))............))------))))))))).......--- ( -26.00, z-score = -1.18, R) >consensus GUGGAUACAGCUGCAACUGGCUGUGUUUUCACAGCCAUUCAGGCACGCCACCGCCUUGCCAAUCGAAGGGCCACACGACCACCCAGAGCCACAUCCACAGC________ ((((........(((..((((((((....))))))))....(((........))).)))...((....))..................))))................. (-22.39 = -22.80 + 0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:54 2011