
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,163,951 – 20,164,033 |
| Length | 82 |
| Max. P | 0.911385 |

| Location | 20,163,951 – 20,164,033 |
|---|---|
| Length | 82 |
| Sequences | 10 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 82.63 |
| Shannon entropy | 0.37239 |
| G+C content | 0.54483 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.99 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20163951 82 - 21146708 AGCGGCGA-CACAUGGCAGCCAGCUGCAAUUAUUUGCAGUU---GGAAGU--UCUGUUUCUGUUGGGAACGC-----GGCGCACAAGGCCAUC .(((.(((-((...(((((((((((((((....))))))))---))....--.)))))..)))))....)))-----(((.......)))... ( -31.70, z-score = -2.05, R) >droWil1.scaffold_180745 1127341 88 - 2843958 AGCAACGAGCACAUGGCAUCCAGCUGCAAUUAUUUGCAGUUAUGGAAAGU--UUCCCCCCUCCCCGGCACCCG---CCAUCCACACCGUUAUC ...((((.....(((((.(((((((((((....)))))))..))))....--......((.....)).....)---))))......))))... ( -22.50, z-score = -2.46, R) >droPer1.super_34 457474 83 + 916997 AGCUGCGA-CACAUGGCAGCCAGCUGCAAUUAUUUGCAGUU---GGAAGU--UCUGUUUCUGUUGGGAACGCG----GGCGCACAAGGCCAUC ........-...(((((..((((((((((....))))))))---))..((--((((((((.....)))))).)----))).......))))). ( -31.00, z-score = -1.60, R) >dp4.chr3 1268136 83 + 19779522 AGCUGCGA-CACAUGGCAGCCAGCUGCAAUUAUUUGCAGUU---GGAAGU--UCUGUUUCUGUUGGGAACGCG----GGCGCACAAGGCCAUC ........-...(((((..((((((((((....))))))))---))..((--((((((((.....)))))).)----))).......))))). ( -31.00, z-score = -1.60, R) >droAna3.scaffold_13266 5636276 83 + 19884421 AGAGGCGA-CACAUGGCAGCCAGCUGCAAUUAUUUGCAGUU---GGAAGU--UCUGUUUCUGUUGGGAACGCG----GGCGCACAAGGCCAUC ...(....-)..(((((..((((((((((....))))))))---))..((--((((((((.....)))))).)----))).......))))). ( -31.20, z-score = -2.16, R) >droEre2.scaffold_4845 21527949 82 - 22589142 AGCGGCGA-CACAUGGCAGCCAGCUGCAAUUAUUUGCAGUU---GGAAGU--UCUGUUUCUGUUGGGAACGC-----GGCGCACAAGGCCAUC .(((.(((-((...(((((((((((((((....))))))))---))....--.)))))..)))))....)))-----(((.......)))... ( -31.70, z-score = -2.05, R) >droYak2.chr2R 20130621 87 - 21139217 AGCGGCGA-CACAUGGCAGCCAGCUGCAAUUAUUUGCAGUU---GGAAGU--UCUGUUUCUGUUGGGAACGCGGCGCGGCGCACAAGGCCAUC .((.(((.-.(((..((..((((((((((....))))))))---))..))--..)))((((....))))))).))..(((.......)))... ( -34.40, z-score = -1.69, R) >droSec1.super_23 43742 82 - 989336 AGCGGCGA-CACAUGGCAGCCAGCUGCAAUUAUUUGCAGUU---GGAAGU--UCUGUUUCUGUUGGGAACGC-----GGCGCACAAGGCCAUC .(((.(((-((...(((((((((((((((....))))))))---))....--.)))))..)))))....)))-----(((.......)))... ( -31.70, z-score = -2.05, R) >droSim1.chr2R_random 2982805 82 - 2996586 AGCGGCGA-CACAUGGCAGCCAGCUGCAAUUAUUUGCAGUU---GGAAGU--UCUGUUUCUGUUGGGAACGC-----GGCGCACAAGGCCAUC .(((.(((-((...(((((((((((((((....))))))))---))....--.)))))..)))))....)))-----(((.......)))... ( -31.70, z-score = -2.05, R) >droGri2.scaffold_15245 797167 77 - 18325388 ------------AUGGCAGCCAGCUGCAAUUAUUUGCAGUUGAGAUUCGCAGUCCAAGGCCAUCAGAAACAGA---CGGCGGCCAAAGUCAU- ------------.((((.(((((((((((....))))))))..(((..((........)).))).........---.))).)))).......- ( -24.70, z-score = -2.15, R) >consensus AGCGGCGA_CACAUGGCAGCCAGCUGCAAUUAUUUGCAGUU___GGAAGU__UCUGUUUCUGUUGGGAACGCG____GGCGCACAAGGCCAUC ............(((((.(((((((((((....))))))))..............(((((.....))))).......))).......))))). (-19.32 = -19.99 + 0.67)

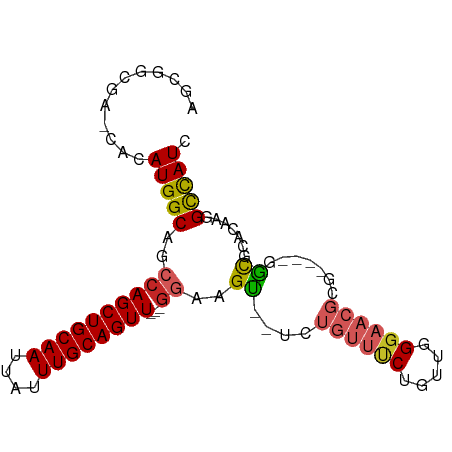
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:51 2011