
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,161,742 – 20,161,834 |
| Length | 92 |
| Max. P | 0.609430 |

| Location | 20,161,742 – 20,161,834 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.01 |
| Shannon entropy | 0.38120 |
| G+C content | 0.45875 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -12.91 |
| Energy contribution | -13.02 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.609430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20161742 92 - 21146708 -------------CGUCGUCGGAAAAGCAGCAUCAGGCGGCAAAUAAUGUAAAACGCCAUUAGGCGAGAUUUUCUCAUACAAACAACAAUUGUGCAUUGGC-ACAC------- -------------.((((((.((.........)).))))))......((((....(((....)))(((.....))).)))).........(((((....))-))).------- ( -23.90, z-score = -2.34, R) >droPer1.super_34 454502 105 + 916997 CCGUCGUCUUCGUCGUAGUAGUCGGGGCAGCAUCAGGCGGCAAAUAAUGUAAAACGCCAUUAGGCGAGAUUUUCUCAUACAAACAACAAUUGUGCAUUGGC-ACAC------- ..(((((((.((.(......).)))))).((.....)))))......((((....(((....)))(((.....))).)))).........(((((....))-))).------- ( -25.80, z-score = -1.09, R) >droAna3.scaffold_13266 5633821 74 + 19884421 -------------------------------AUCAGGCGGCAAAUAAUGUAAAACGCCAUUAGGCGAGAUUUUCUCAUACAAACAACAAUUGUGCAUUGGC-ACAC------- -------------------------------................((((....(((....)))(((.....))).)))).........(((((....))-))).------- ( -15.60, z-score = -1.61, R) >droSec1.super_23 41376 92 - 989336 -------------CGUCGUCGGAAGAGCGGCAUCAGGCGGCAAAUAAUGUAAAACGCCAUUAGGCGAGAUUUUCUCAUACAAACAACAAUUGUGCAUUGGC-ACAC------- -------------.((((((.((.(.....).)).))))))......((((....(((....)))(((.....))).)))).........(((((....))-))).------- ( -24.60, z-score = -2.03, R) >droSim1.chr2R_random 2980439 92 - 2996586 -------------CGUCGUCGGAAGAGCAGCAUCAGGCGGCAAAUAAUGUAAAACGCCAUUAGGCGAGAUUUUCUCAUACAAACAACAAUUGUGCAUUGGC-ACAC------- -------------.((((((.((.(.....).)).))))))......((((....(((....)))(((.....))).)))).........(((((....))-))).------- ( -24.60, z-score = -2.37, R) >droWil1.scaffold_180745 1118396 86 - 2843958 ------------------------GGCCAGCAUCAGGCGGCAAAUAAUGUAAAACGCCAUUAGGCGAGAUUUUCUCAUACAAACAACAAUUGUGCAUUGGCAACGUCUAC--- ------------------------..........(((((.....(((((((....(((....)))(((.....)))................)))))))....)))))..--- ( -19.70, z-score = -1.24, R) >droVir3.scaffold_12875 7250041 88 - 20611582 ------------------------CGCCGGCAUCAGGCGGCAAAUAAUGUAAAACGCCAUUAGGCGAGAUUUUCUCAUACAAACAACAAUUGUGCAUUGGC-ACACAGAGCGC ------------------------.((((.(....).))))......((((....(((....)))(((.....))).)))).........(((((....))-)))........ ( -24.70, z-score = -2.12, R) >droMoj3.scaffold_6496 26005339 91 - 26866924 ---------------------CGUCGUCGGCAUCAGGCGGCAAAUAAUGUAAAACGCCAUUAGGCGAGAUUUUCUCAUACAAACAACAAUUGGGCAUUGGC-ACACAGGGCGC ---------------------((((((((.(....).))))...((((((.....(((....)))(((.....))).................))))))..-......)))). ( -22.10, z-score = -1.42, R) >droGri2.scaffold_15245 793962 97 - 18325388 ---------------CGACUUCGACGUCGGCAUCAGGCGGCAAAUAAUGUAAAACGCCAUUAGGCGAGAUUUUCUCGUACAAACAACAAUUGUGCAUUGGC-AUACAGCGAAC ---------------....((((.(((((.(....).))))......((((....((((......(((.....)))((((((.......))))))..))))-.))))))))). ( -23.90, z-score = -1.13, R) >consensus _______________________AGGGCAGCAUCAGGCGGCAAAUAAUGUAAAACGCCAUUAGGCGAGAUUUUCUCAUACAAACAACAAUUGUGCAUUGGC_ACAC_______ ................................((((...(((.....(((.....(((....)))(((.....))).........)))....))).))))............. (-12.91 = -13.02 + 0.11)

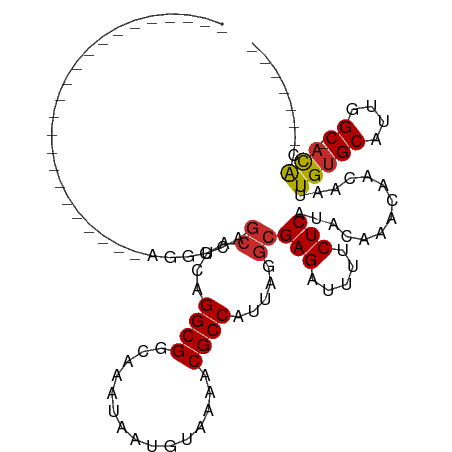
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:50 2011