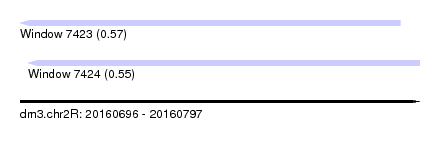
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,160,696 – 20,160,797 |
| Length | 101 |
| Max. P | 0.574242 |

| Location | 20,160,696 – 20,160,792 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 97.74 |
| Shannon entropy | 0.03577 |
| G+C content | 0.59896 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -34.11 |
| Energy contribution | -34.17 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.574242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
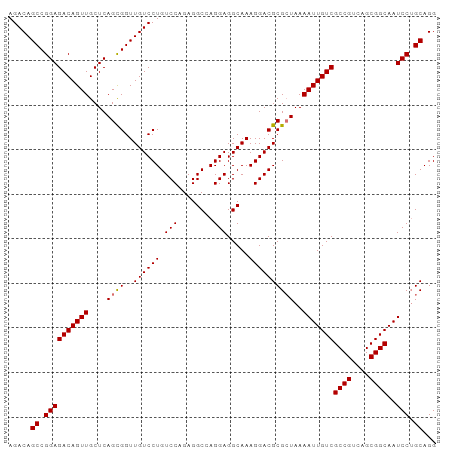
>dm3.chr2R 20160696 96 - 21146708 AGACAGCCGGAGACAGUUGCUCAGCGGUUGUCCUGUCCAGAGGCCAGGAGGCAAAGGACGCGCUAAAAUUGUCGCCGUCAGCGGCAAUCCUGCAGG .....((.((((((((((....((((..((((((........(((....)))..))))))))))..)))))))((((....))))..))).))... ( -37.10, z-score = -1.36, R) >droSec1.super_23 40354 96 - 989336 AGACAGCCGGAGACAGUUGCUCAGUGGUUGUCCUGUCCAGAGGCCAGGAGGCAAAGGACGCGAUAAAAUUGUCGCCGUCAGCGGCAAUCCUGCAGG ...((((.(....).))))(((((.(((((((((((((....(((....)))...))))((((((....))))))....)).)))))))))).)). ( -35.80, z-score = -1.69, R) >droYak2.chr2R 20127117 96 - 21139217 AGACAGCCGGAGACAGUUGCUCAGCGGUUGUCCUGUCCAGAGGCGAGGAGGCAAAGGACGCGCUAAAAUUGUCGCCGUCAGCGGCAAUCCAGCAGG .....((.((((((((((....((((..((((((.(((........))).....))))))))))..)))))))((((....))))..))).))... ( -35.80, z-score = -1.30, R) >droEre2.scaffold_4845 21524515 96 - 22589142 AGACAGCCGGAGACAGUUGCUCAGCGGUUGUCCUGUCCAGAGGCGAGGAGGCAAAGGACGCGCUAAAAUUGUCGCCGUCAGCGGCAAUCCUGCAGG .....((.((((((((((....((((..((((((.(((........))).....))))))))))..)))))))((((....))))..))).))... ( -35.80, z-score = -0.87, R) >consensus AGACAGCCGGAGACAGUUGCUCAGCGGUUGUCCUGUCCAGAGGCCAGGAGGCAAAGGACGCGCUAAAAUUGUCGCCGUCAGCGGCAAUCCUGCAGG .....((.((((((((((....((((..((((((.(((........))).....))))))))))..)))))))((((....))))..))).))... (-34.11 = -34.17 + 0.06)


| Location | 20,160,698 – 20,160,797 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 96.97 |
| Shannon entropy | 0.04984 |
| G+C content | 0.58478 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -34.11 |
| Energy contribution | -34.17 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
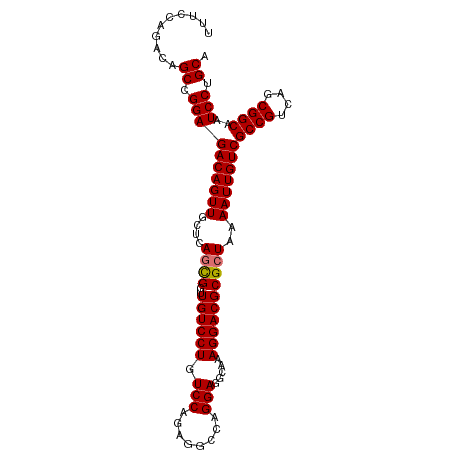
>dm3.chr2R 20160698 99 - 21146708 CUUCCAGACAGCCGGAGACAGUUGCUCAGCGGUUGUCCUGUCCAGAGGCCAGGAGGCAAAGGACGCGCUAAAAUUGUCGCCGUCAGCGGCAAUCCUGCA ..........((.((((((((((....((((..((((((........(((....)))..))))))))))..)))))))((((....))))..))).)). ( -37.10, z-score = -1.19, R) >droSec1.super_23 40356 98 - 989336 -UUCCAGACAGCCGGAGACAGUUGCUCAGUGGUUGUCCUGUCCAGAGGCCAGGAGGCAAAGGACGCGAUAAAAUUGUCGCCGUCAGCGGCAAUCCUGCA -.....(((((((.(((.(....))))...)))))))..((((....(((....)))...))))((((((....))))((((....))))......)). ( -35.10, z-score = -1.40, R) >droYak2.chr2R 20127119 99 - 21139217 UUUCCAGACAGCCGGAGACAGUUGCUCAGCGGUUGUCCUGUCCAGAGGCGAGGAGGCAAAGGACGCGCUAAAAUUGUCGCCGUCAGCGGCAAUCCAGCA ..........((.((((((((((....((((..((((((.(((........))).....))))))))))..)))))))((((....))))..))).)). ( -35.80, z-score = -1.20, R) >droEre2.scaffold_4845 21524517 99 - 22589142 UUUCCAGACAGCCGGAGACAGUUGCUCAGCGGUUGUCCUGUCCAGAGGCGAGGAGGCAAAGGACGCGCUAAAAUUGUCGCCGUCAGCGGCAAUCCUGCA ..........((.((((((((((....((((..((((((.(((........))).....))))))))))..)))))))((((....))))..))).)). ( -35.80, z-score = -0.83, R) >consensus UUUCCAGACAGCCGGAGACAGUUGCUCAGCGGUUGUCCUGUCCAGAGGCCAGGAGGCAAAGGACGCGCUAAAAUUGUCGCCGUCAGCGGCAAUCCUGCA ..........((.((((((((((....((((..((((((.(((........))).....))))))))))..)))))))((((....))))..))).)). (-34.11 = -34.17 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:49 2011