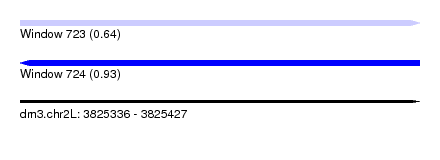
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,825,336 – 3,825,427 |
| Length | 91 |
| Max. P | 0.929427 |

| Location | 3,825,336 – 3,825,427 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.89 |
| Shannon entropy | 0.40552 |
| G+C content | 0.43080 |
| Mean single sequence MFE | -23.02 |
| Consensus MFE | -15.98 |
| Energy contribution | -15.39 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3825336 91 + 23011544 -------------AAAUACCGACCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAAGCA-GCAAAAGACUGCAAAUAAACAUCCACAAGUUCCCAAACA------------ -------------.............((((((..(((((((..(((...........)))..(((-((....).))))......))))))).......)))))).------------ ( -19.60, z-score = -1.74, R) >droAna3.scaffold_12916 1831433 102 + 16180835 --------------AAGACCGACCCUGUUUGGAUUGGGUGUAAUGGAUAUUUUGACGCCGGAGCA-GCAAAAGACUGCAAAUAAACAUCCACACGUUCCCAAACACACGGUCCGCUU --------------..(((((....(((((((...(..(((..(((((..((((....))))(((-((....).))))........))))).)))..))))))))..)))))..... ( -28.20, z-score = -1.43, R) >droEre2.scaffold_4929 3876657 90 + 26641161 --------------AAGACCGACCCUGUUUGGAUUGGGUGUAAUGGAUAUUUUGACGCCGAAGCA-GCGAAAGACUGCAAAUAAACAUCCACAAGUUCCCAAACA------------ --------------....((((......)))).(((((.....(((((..((((....))))(((-((....).))))........)))))......)))))...------------ ( -23.00, z-score = -1.86, R) >droYak2.chr2L 3829193 90 + 22324452 --------------AAGAGCGACCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAAGCA-GCAAAAGACUGCAAAUAAACAUCCACAAGUUCCCAAACA------------ --------------............((((((..(((((((..(((...........)))..(((-((....).))))......))))))).......)))))).------------ ( -19.60, z-score = -1.26, R) >droSec1.super_5 1927174 91 + 5866729 -------------AAAUACCGACCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAAGCA-GCAAAAGACUGCAAAUAAACAUCCACAAGUUCCCAAACA------------ -------------.............((((((..(((((((..(((...........)))..(((-((....).))))......))))))).......)))))).------------ ( -19.60, z-score = -1.74, R) >droSim1.chr2L 3782240 91 + 22036055 -------------AAAUAGCGACCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAAGCA-GCAAAAGACUGCAAAUAAACAUCCACAAGUUCCCAAACA------------ -------------.............((((((..(((((((..(((...........)))..(((-((....).))))......))))))).......)))))).------------ ( -19.60, z-score = -1.43, R) >droWil1.scaffold_180708 9704639 107 + 12563649 --UAAAGAGAGGGAGCCAAAAACUGUGUUUGGAUUGGAUGUAAUAGAUAUUUUGACGCUGAAGCA-ACUAAAAACGGCAAAUAAACAUCCACAAAAUCCAAAAUAAAAAU------- --.........................((((((((((((((....((....))...((((.....-........))))......)))))))....)))))))........------- ( -16.22, z-score = -0.79, R) >droVir3.scaffold_13246 2386134 104 + 2672878 CAAGCUGAGCGGCGCA-GCAACCCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAGCCAAGCAAAGGGCUGCAAAUAAACAUCCACAAGUUCCAAAACA------------ ...(((....)))(((-((..((..(((((((.((((...(((........)))...)))).)))))))..)))))))...........................------------ ( -28.00, z-score = -0.46, R) >droMoj3.scaffold_6500 26472281 105 + 32352404 CAAGAAGAGAGCGGUGUGCAACCCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAGCCAAGCAAAGGGCUGCAAAUAAACAUCCACAAGUUCCAAAACA------------ ......(.((((.(((((((.(((.(((((((.((((...(((........)))...)))).)))))))..))).))))..........)))..)))))......------------ ( -27.40, z-score = -0.93, R) >droGri2.scaffold_15252 5842800 105 + 17193109 CAAGGGGCGAGUGGCGCACAACCCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAUGCAUGCAAAGGGCUGCAAAUAAACAUCCACAAGUUCCAAAACA------------ ..(((((...(((...)))..))))).((((((.(((((((..(((...........))).((((.((.....)))))).....))))))).....))))))...------------ ( -29.00, z-score = -0.27, R) >consensus _____________AAAUACCGACCCUGUUUGGAUUGGAUGUAAUGGAUAUUUUGACGCCGAAGCA_GCAAAAGACUGCAAAUAAACAUCCACAAGUUCCCAAACA____________ .........................((((((((.(((((((..(((...........)))......(((......)))......))))))).....))).)))))............ (-15.98 = -15.39 + -0.59)

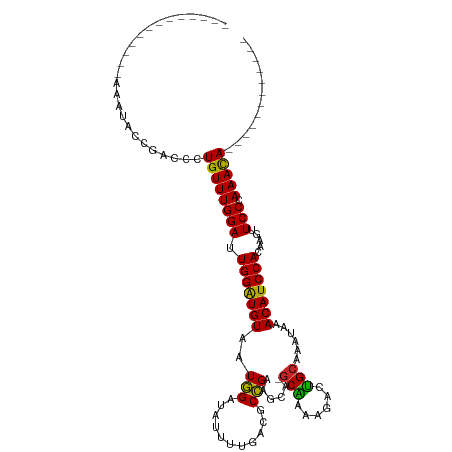
| Location | 3,825,336 – 3,825,427 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.89 |
| Shannon entropy | 0.40552 |
| G+C content | 0.43080 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -16.75 |
| Energy contribution | -17.37 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3825336 91 - 23011544 ------------UGUUUGGGAACUUGUGGAUGUUUAUUUGCAGUCUUUUGC-UGCUUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGGUCGGUAUUU------------- ------------((((((((.....((((((((((....((((....))))-.((....))....)))))))))).........))))))))............------------- ( -25.24, z-score = -2.20, R) >droAna3.scaffold_12916 1831433 102 - 16180835 AAGCGGACCGUGUGUUUGGGAACGUGUGGAUGUUUAUUUGCAGUCUUUUGC-UGCUCCGGCGUCAAAAUAUCCAUUACACCCAAUCCAAACAGGGUCGGUCUU-------------- ..((.((((.(((..(((((.....((((((((((....((((....))))-.((....))....))))))))))....))))).....))).)))).))...-------------- ( -34.90, z-score = -2.45, R) >droEre2.scaffold_4929 3876657 90 - 26641161 ------------UGUUUGGGAACUUGUGGAUGUUUAUUUGCAGUCUUUCGC-UGCUUCGGCGUCAAAAUAUCCAUUACACCCAAUCCAAACAGGGUCGGUCUU-------------- ------------....((((.....((((((((((....((........))-.((....))....))))))))))....))))((((.....)))).......-------------- ( -23.50, z-score = -1.27, R) >droYak2.chr2L 3829193 90 - 22324452 ------------UGUUUGGGAACUUGUGGAUGUUUAUUUGCAGUCUUUUGC-UGCUUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGGUCGCUCUU-------------- ------------((((((((.....((((((((((....((((....))))-.((....))....)))))))))).........))))))))...........-------------- ( -25.24, z-score = -2.39, R) >droSec1.super_5 1927174 91 - 5866729 ------------UGUUUGGGAACUUGUGGAUGUUUAUUUGCAGUCUUUUGC-UGCUUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGGUCGGUAUUU------------- ------------((((((((.....((((((((((....((((....))))-.((....))....)))))))))).........))))))))............------------- ( -25.24, z-score = -2.20, R) >droSim1.chr2L 3782240 91 - 22036055 ------------UGUUUGGGAACUUGUGGAUGUUUAUUUGCAGUCUUUUGC-UGCUUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGGUCGCUAUUU------------- ------------((((((((.....((((((((((....((((....))))-.((....))....)))))))))).........))))))))............------------- ( -25.24, z-score = -2.33, R) >droWil1.scaffold_180708 9704639 107 - 12563649 -------AUUUUUAUUUUGGAUUUUGUGGAUGUUUAUUUGCCGUUUUUAGU-UGCUUCAGCGUCAAAAUAUCUAUUACAUCCAAUCCAAACACAGUUUUUGGCUCCCUCUCUUUA-- -------.........((((((...((((((((((...(((.((.......-.))....)))...))))))))))...)))))).(((((.......))))).............-- ( -18.10, z-score = -1.59, R) >droVir3.scaffold_13246 2386134 104 - 2672878 ------------UGUUUUGGAACUUGUGGAUGUUUAUUUGCAGCCCUUUGCUUGGCUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGGGUUGC-UGCGCCGCUCAGCUUG ------------((..(((((....((((((((((...(((((((........))))..)))...))))))))))....)))))..))....((.((...-.)).)).......... ( -30.20, z-score = -0.75, R) >droMoj3.scaffold_6500 26472281 105 - 32352404 ------------UGUUUUGGAACUUGUGGAUGUUUAUUUGCAGCCCUUUGCUUGGCUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGGGUUGCACACCGCUCUCUUCUUG ------------((..(((((....((((((((((...(((((((........))))..)))...))))))))))....)))))..))...(((((..((.....))...))))).. ( -30.10, z-score = -2.14, R) >droGri2.scaffold_15252 5842800 105 - 17193109 ------------UGUUUUGGAACUUGUGGAUGUUUAUUUGCAGCCCUUUGCAUGCAUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGGGUUGUGCGCCACUCGCCCCUUG ------------((..(((((....((((((((((....((((....))))((((....))))..))))))))))....)))))..))...((((((.(((...)))..)))))).. ( -33.50, z-score = -2.31, R) >consensus ____________UGUUUGGGAACUUGUGGAUGUUUAUUUGCAGUCUUUUGC_UGCUUCGGCGUCAAAAUAUCCAUUACAUCCAAUCCAAACAGGGUCGGUAUUU_____________ ..................(((....((((((((((....((((....))))..((....))....))))))))))....)))..(((.....)))...................... (-16.75 = -17.37 + 0.62)
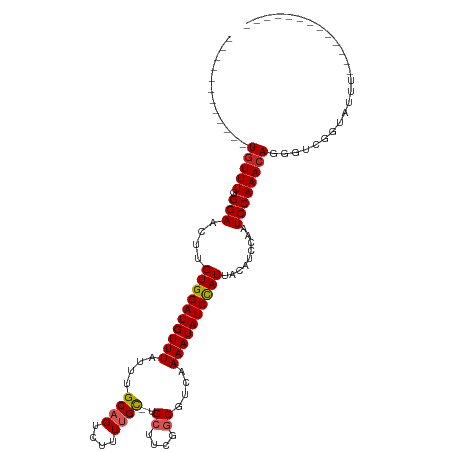
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:44 2011