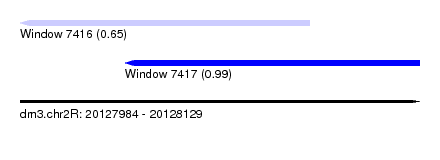
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,127,984 – 20,128,129 |
| Length | 145 |
| Max. P | 0.985003 |

| Location | 20,127,984 – 20,128,089 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.51 |
| Shannon entropy | 0.52184 |
| G+C content | 0.54930 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -18.52 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20127984 105 - 21146708 ------AGCUGGGAGCUGGGAGCUAAG---GGAGCAGGGGAC-UGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUGU-AAAGGAAUCGUUUAAAAUAAAUGACAGGCGGACCCCC- ------.(((...(((.....)))...---..))).((((.(-((.(...(((((.(((....))).)))))......-.......(((((((...)))))))..).))).)))).- ( -37.30, z-score = -1.64, R) >droSim1.chr2R 18610584 98 - 19596830 -------------AGCUGGGAGCUAAG---GGAGCAGGGGAC-CGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUGU-AAAGGAAUCGUUUAAAAUAAAUGACAGGCGAGCCCCC- -------------........(((...---..))).((((..-((.(...(((((.(((....))).)))))......-.......(((((((...)))))))..).))..)))).- ( -34.20, z-score = -1.30, R) >droYak2.chr2R 20093660 115 - 21139217 GCAAGUGGCAGGGGGUAGGGGCAUGGGGCAGGGGCAGGGGAC-UGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUGU-AAAGGAAUCGUUUAAAAUAAAUGACAGGCGGGCCCCCC ((.....)).((((((....((.((..(((.((.(((....)-)).)...(((((.(((....))).))))).).)))-.......(((((((...))))))))).))..)))))). ( -44.90, z-score = -1.89, R) >droWil1.scaffold_180701 3683641 108 + 3904529 -----UGUGUGUGGAGGGGGGAAUGGGACGUCAGCGGUGG---CGGCAACGUUGGCGGCGACAGCCUCCGACAUGUGUAAAAGGAAUCGUUUAAAAUAAAUGACAGCAGCGCCCCC- -----..........(((((........(((((....)))---))((...(((((.(((....))).)))))..............(((((((...)))))))..))....)))))- ( -33.50, z-score = 0.32, R) >droPer1.super_34 422534 109 + 916997 ---GGCAUCAGAAAGGACAACAGCAAAACAACAAUAAAUGCG---GCAACGUUGGCGGCGACAGACUCCGACAUGUGU-AAAGGAAUCGUUUAAAAUAAAUGACAGCGGCGCCCCC- ---(((...........((((.(((.............)))(---....)))))((.((.......(((.((....))-...))).(((((((...)))))))..)).)))))...- ( -24.42, z-score = -0.22, R) >droAna3.scaffold_13266 5604110 101 + 19884421 ------------AACAUCAACAGC--AACAACAAUAAAUGCGGCAGCAACGUUGGCGGCGAAAGCCUCCGACAUGUGU-AAAGGAAUCGUUUAAAAUAAAUGACAGGCGAGCCCCC- ------------..........((--(...........)))(((.((...(((((.(((....))).)))))......-.......(((((((...)))))))...))..)))...- ( -27.30, z-score = -1.85, R) >consensus ________C_G_GAGCUGGGAAAUAAGACAGCAGCAGGGGAC_CGGCAACGUUGGCGGCGAAAGCCUCCGACAUGUGU_AAAGGAAUCGUUUAAAAUAAAUGACAGGCGAGCCCCC_ ............................................(((...(((((.(((....))).)))))..............(((((((...))))))).......))).... (-18.52 = -19.18 + 0.67)


| Location | 20,128,022 – 20,128,129 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.57 |
| Shannon entropy | 0.53934 |
| G+C content | 0.57954 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -21.94 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985003 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 20128022 107 - 21146708 AGCCGAGCUGUACUACUAUAUAGCUGAAACUGAAGCUG------GAAGCUGGGAGCUGGGAGCUAAG---GGAGCAGGGGACUGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUG .(((.(((((((......)))))))(...((..((((.------..(((.....)))...))))..)---)...)((....)))))...(((((.(((....))).)))))..... ( -40.00, z-score = -2.00, R) >droSim1.chr2R 18610622 100 - 19596830 AGCCGAGCUGUACUACUAUAUAGCUGAAAACGAA-------------GCUGGGAGCUGGGAGCUAAG---GGAGCAGGGGACCGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUG .((((((((...(((((...(((((........)-------------))))..)).))).))))...---......(....)))))...(((((.(((....))).)))))..... ( -37.50, z-score = -1.89, R) >droYak2.chr2R 20093699 113 - 21139217 AGCCGCGC---AGUACUAUAUAGCUGAAACUGAAGCUGUGGCAAGUGGCAGGGGGUAGGGGCAUGGGGCAGGGGCAGGGGACUGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUG .(((.(((---..((((.(((((((........)))))))((.....))....))))...))..).)))...(.(((....))).)...(((((.(((....))).)))))..... ( -43.20, z-score = -1.25, R) >droAna3.scaffold_13266 5604148 98 + 19884421 -----AGCUAUAUGGCUAUAUAGCCCUGCCUGCC-------------ACCAGCAACAUCAACAGCAACAACAAUAAAUGCGGCAGCAACGUUGGCGGCGAAAGCCUCCGACAUGUG -----.(((((((....))))))).((((((((.-------------....))).........(((...........))))))))....(((((.(((....))).)))))..... ( -33.30, z-score = -2.48, R) >consensus AGCCGAGCUGUACUACUAUAUAGCUGAAACUGAA_____________GCUGGGAGCAGGGACAUAAG___GGAGCAGGGGACCGGCGACGUUGGCGGCGAAAGCCUCCGACAUGUG ......((((((......))))))...........................(..(((((......................)))))..)(((((.(((....))).)))))..... (-21.94 = -22.88 + 0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:43 2011