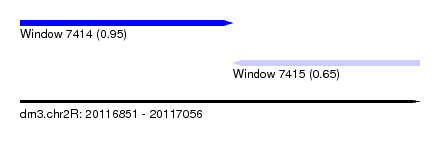
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,116,851 – 20,117,056 |
| Length | 205 |
| Max. P | 0.952954 |

| Location | 20,116,851 – 20,116,960 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 67.80 |
| Shannon entropy | 0.56605 |
| G+C content | 0.60501 |
| Mean single sequence MFE | -45.82 |
| Consensus MFE | -24.76 |
| Energy contribution | -26.52 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.952954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
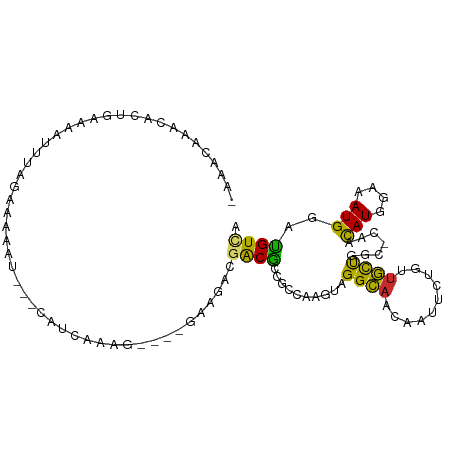
>dm3.chr2R 20116851 109 + 21146708 ---GCGGCCGGAGUCCUGGUGGCUCCAACUCCGGCUCCGGCUUCGGCUGCAUUGUUGUUGGAGCAGCGGGCGGAUAUGGUCAUUAGGUUGCCAUGGCGGCCGGUCGGGCGAU ---(((((((((((..(((.....))))))))))))(((((..(((((((.(((((((....))))))).....(((((.((......)))))))))))))))))))))... ( -52.90, z-score = -1.40, R) >droSec1.super_5 356135 109 + 5866729 ---GCGGCCGGAGUCCUGGUGGCUCCAACUCCGGCUCCGGCUUCGGCUGCAUUGUUGUUGGAGCAGCGGGCGGAUAUGGUCAUUAGGUUGCCAUGGCGGCCGGUCGGGCGAU ---(((((((((((..(((.....))))))))))))(((((..(((((((.(((((((....))))))).....(((((.((......)))))))))))))))))))))... ( -52.90, z-score = -1.40, R) >droYak2.chr2R 20081328 100 + 21139217 ------------GCGGCUCCAGCUCCAGCCGCUGCUCCAAUUCCGGUUGCAUUGUUGUUGGAGCAGCGGGCGGAUAUGGUCAUUAGGUUGCCAUGGCGGCCGGUCGGGCGGU ------------.((((((((..(((..(((((((((((((..((((...))))..)))))))))))))..)))..)))......((((((....)))))))))))...... ( -49.80, z-score = -2.37, R) >droEre2.scaffold_4845 21479410 100 + 22589142 ------------GCGGCUCCAACUCCUGCUCCAGCUCCAACUCCGGCUGCAUUGUUGUUGGAGCAGCGGGCGGAUAUGGUCAUUAGGUUGCCAUGGCGGCCGGUCGGGCGAU ------------.((((((((..(((.(((((.((((((((..(((.....)))..)))))))).).)))))))..)))......((((((....)))))))))))...... ( -41.00, z-score = -0.25, R) >droWil1.scaffold_180701 3662501 106 - 3904529 GUCGUUGUUGGUAAUGUGGAAGCUGGCGGGCAAGUUGUGGGUGUUCCAUUGUUAUUAUUGGAGCAACGUAAUGAAAUGGUCAUUAGAUUGC--UAGCAUCAUGACGAU---- (((((((((((((((.(((....((((.(....((((((..(((((((.((....)).))))))).))))))....).))))))).)))))--)))))..)))))...---- ( -32.50, z-score = -2.36, R) >consensus ___G__G__GG_GCCGUGGCAGCUCCAGCUCCAGCUCCGGCUCCGGCUGCAUUGUUGUUGGAGCAGCGGGCGGAUAUGGUCAUUAGGUUGCCAUGGCGGCCGGUCGGGCGAU ............(((......((((((((..(((...((((....))))..)))..))))))))..(((.(((.(((((.((......)))))))....))).))))))... (-24.76 = -26.52 + 1.76)


| Location | 20,116,960 – 20,117,056 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 60.49 |
| Shannon entropy | 0.79064 |
| G+C content | 0.45362 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -7.12 |
| Energy contribution | -6.10 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20116960 96 - 21146708 -AAACAAACACUGGAAAUCGAGCGAAAU---CAACAAAG----GAAGUCGACGCCGCCCUGUAGGCAACGAUUCCGCUGCUAGC-CAACAUGGAAAUGGACGUCA -..........(((......((((....---)......(----(((.(((..(((........)))..)))))))))).....)-)).(((....)))....... ( -20.10, z-score = 0.51, R) >droSec1.super_5 356244 96 - 5866729 -AAACAGGCACUGGAAAUCGAGCGAAAU---CAACAAAG----GAAGACGACGCCGCCCUGUAGGCAACGAUUCCGCUGCUAGC-CAACAUGGAAAUGGACGUCA -.....(((..(((......((((....---)......(----(((..((..(((........)))..)).))))))).....)-)).(((....)))...))). ( -21.90, z-score = 0.32, R) >droYak2.chr2R 20081428 96 - 21139217 -AAACAAACACUGGAAAUCGAGCAAAAU---CAACAAGG----GAAGACGACGCCGCCCUGUAGGCAACGAUUCCGCUGCUAGC-CAACAUGGAAAUGGACGUCA -..........(((......((((....---(.....)(----(((..((..(((........)))..)).))))..))))..)-)).(((....)))....... ( -17.60, z-score = 1.06, R) >droAna3.scaffold_13266 5595510 94 + 19884421 -AAACAAACUCUGCAAAAUUGAUAAAAU---CCUUGAA------AGGACGACGCCGCCGAGUAGGCAACCAAUUUGCUGCUAGC-CAUCAUGGAAAUGGACGUUA -..........................(---(((....------)))).(((((((((.....)))..(((....((.....))-.....)))....)).)))). ( -17.40, z-score = 0.27, R) >dp4.chr3 1223847 98 + 19779522 -AAACCAAC-UGAAAUAUUUCUAGAAAUAGGCGUCCACA----GCGGAAGACGCUGAGAAGCAGGCAACAGUUCUGUUGCCGGC-UAACAUGGAAAUGGAUGUUA -........-..........(((....)))(((((((((----(((.....)))))...(((.(((((((....))))))).))-)..........))))))).. ( -34.70, z-score = -4.18, R) >droPer1.super_34 413397 98 + 916997 -AAACCAAC-UGAAAAAUUUGGAGAAAUAGGCGUCCACA----GCGGAAGACGCUGAGAAGCAGGCAACAGUUCUGUUGCCGGC-UAACAUGGAAAUGGAUGUUA -...((((.-(.....).)))).......((((((((((----(((.....)))))...(((.(((((((....))))))).))-)..........)))))))). ( -35.40, z-score = -4.45, R) >droWil1.scaffold_180701 3662607 105 + 3904529 AAACCAAAUAGAAAUUUGUCAAAAAGAUUCCAUUCAACGCUCUUAAGUAGGCAACAGUUUUUUGGUAUUCUCUUACUCACUGGCUCAGUAUGGAAAUGGAUGUCA ....(((((....))))).......((((((((((.((((.((.....))))..((((.....((((......)))).)))).....))...).)))))).))). ( -15.20, z-score = 1.05, R) >droGri2.scaffold_15245 739614 86 - 18325388 -----GAGCAACGAAAAUAUAAUAGA-----CAUCAA-------CACAGCGCGCAGCAAAGUAGGCAACAGUUUU-UUGUUGCG-CACCAUGGAAAUGGAUGUCA -----...................((-----((((..-------......(((((((((((..(....)....))-))))))))-)..(((....))))))))). ( -28.50, z-score = -4.45, R) >droVir3.scaffold_12875 7194120 92 - 20611582 -AAACAGGCAAGAAAAAAACAUUUCG---CAAAUCAA-------CACCGCGCGCAGGAAAGUAGGCAACAGUUUU-UUGUUGCG-CACCAUGGAAAUGGAUGUCA -.....((((.((((......)))).---........-------....((((((((((((.(.(....)).))))-))).))))-).((((....)))).)))). ( -25.60, z-score = -2.11, R) >consensus _AAACAAACACUGAAAAUUUAGAAAAAU___CAUCAAAG____GAAGACGACGCCGCCAAGUAGGCAACAAUUCUGUUGCUGGC_CAACAUGGAAAUGGAUGUCA .................................................((((..........((((..........)))).......(((....)))..)))). ( -7.12 = -6.10 + -1.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:42 2011