| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,116,717 – 20,116,816 |
| Length | 99 |
| Max. P | 0.607332 |

| Location | 20,116,717 – 20,116,816 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.50 |
| Shannon entropy | 0.65338 |
| G+C content | 0.60394 |
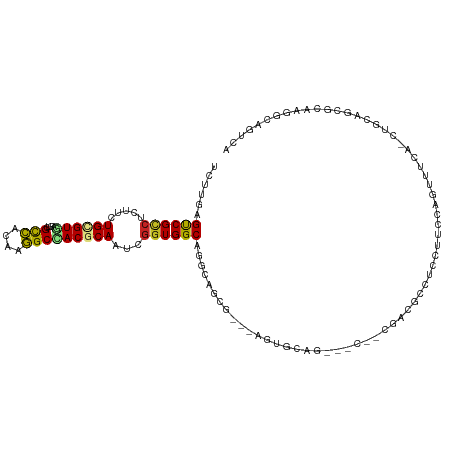
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -16.24 |
| Energy contribution | -15.99 |
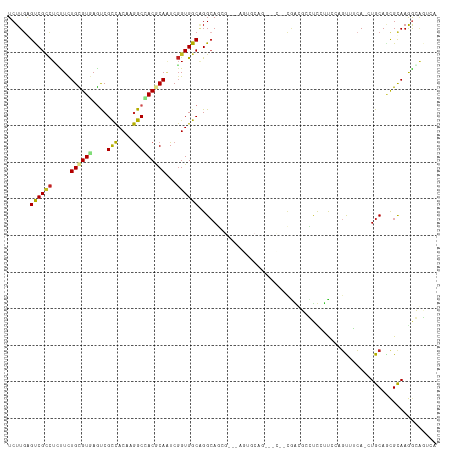
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.607332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
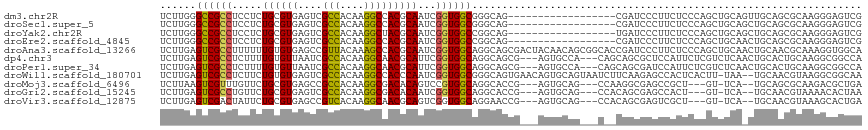
>dm3.chr2R 20116717 99 - 21146708 UCUUGGGCCGCCUCCUCUGCGUGAGUCGCCACAAGGCCACGCAAUCGGUGGCGGGCAG------------------CGAUCCCUUCUCCCAGCUGCAGUUGCAGCGCAAGGGAGUCG .....(.(((((.((..((((((.(((.......)))))))))...)).))))).)..------------------(((((((((......(((((....)))))..)))))).))) ( -46.40, z-score = -1.98, R) >droSec1.super_5 356001 99 - 5866729 UCUUGGGCCGCCUCCUCUGCGUGAGUCGCCACAAGGCCACGCAAUCGGUGGCGGGCAG------------------CGAUCCCUUCUCCCAGCUGCAGCUGCAGCGCAAGGGAGUCG .....(.(((((.((..((((((.(((.......)))))))))...)).))))).)..------------------(((((((((......(((((....)))))..)))))).))) ( -46.40, z-score = -1.60, R) >droYak2.chr2R 20081194 99 - 21139217 UCUUGGGCCGCCUCCUCUGCGUGAGUCGCCACAAGGCUACGCAAUCGGUGGCCGGCAG------------------UGAUCCCUUCUCCCAGCUGCAGCUGCAGCGCAAGGGAGUCG .....(((((((.....(((((.((((.......)))))))))...)))))))((.(.------------------...).))..(((((.(((((....)))))....)))))... ( -42.40, z-score = -1.02, R) >droEre2.scaffold_4845 21479276 99 - 22589142 UCUUGGGCCGCCUCCUCUGCGUGAGUCGCCACAAGGCCACGCAAUCGGUGGCCGGCAG------------------CGAUCCCUUCUCCCAGCUGCAACUGCAGCGCAAGGGAGUCG .....(((((((.....((((((.(((.......)))))))))...))))))).....------------------(((((((((......(((((....)))))..)))))).))) ( -44.50, z-score = -1.92, R) >droAna3.scaffold_13266 5595243 117 + 19884421 UCUUGAGUCGCCUUUUUUGUGUGAGCCGUUACAAAGCCACGCAAUCGGUGGCAGGCAGCGACUACAACAGCGGCACCGAUCCCUUCUCCCAGCUGCAACUGCAACGCAAAGGUGGCA ......((((((((...(((((..((.(((.....(((((.......)))))..(((((...........((....)).............)))))))).)).))))))))))))). ( -39.05, z-score = -0.49, R) >dp4.chr3 1223580 111 + 19779522 UCUUGAGUCGCCUCUUUUGUGUUAAUCGCCACAAGGCAACGCAUUCGGUGGCAGGCAGCG---AGUGCCA---CAGCAGCGCUCCAUUCUCGUCUCAACUGCACUGCAAGGCGGCCA ...((.(((((((....((((......(((....)))...(((((((.((.....)).))---)))))))---))((((.((..................)).)))).))))))))) ( -40.07, z-score = -1.18, R) >droPer1.super_34 413130 111 + 916997 UCUUGAGUCGCCUCUUUUGUGUUAAUCGCCACAAGGCAACGCAUUCGGUGGCAGGCAGCG---AGUGCCA---CAGCAGCGAUCCAUUCUCGUCUCAACUGCACUGCAAGGCGGCCA ...((.(((((((....((((((....(((....))))))))).......((((((((.(---(((((..---..)))((((.......)))))))..)))).)))).))))))))) ( -37.40, z-score = -0.60, R) >droWil1.scaffold_180701 3662325 114 + 3904529 UCUUGAGUCGCCUCUUCUGUGUGAGUCGCCACAAGGCCACCCAAUCGGUGGCGGGCAGUGAACAGUGCAGUAAUCUUCAAGAGCCACUCACUU-UAA--UGCAACGUAAGGCGGCAA ......(((((((....(((((((((.(((.....((((((.....))))))..(((.(....).)))............).)).))))))..-...--.))).....))))))).. ( -37.10, z-score = -0.94, R) >droMoj3.scaffold_6496 25934027 105 - 26866924 UCUUAAGUCGUUUGUUCUGCGUGAGCCGCCACAAGGCGACACAGUCCGUGGCAGGCACCG---AGUGCAG---CCAAGGCGAGCCGCU---GU-UCA--UGCAGCGCAAGACGCUGA .....((.(((((((.((((((((((((((....)))).........(((((..((((..---.)))).(---(....))..))))).---))-)))--))))).)).))))))).. ( -44.70, z-score = -1.68, R) >droGri2.scaffold_15245 739353 105 - 18325388 UCUUGAGUCGCCUGUUCUGCGUGAGUCGCCACAAGGCGACACAAUCGGUGGCAGGCACCG---AGUGCAG---CCACAGCGAGCCACU---GU-UCA--UGCAACGUAAAACACUAA .((((.((.((((((((((..((.((((((....)))))).))..))).)))))))))))---))(((((---(....))((((....---))-)).--)))).............. ( -37.70, z-score = -1.36, R) >droVir3.scaffold_12875 7193877 105 - 20611582 UCUUGAGUCGACUAUUCUGCGUGAGCCGUCACAAGGCAACGCAGUCGGUGGCAGGAACCG---AGUGCAG---CCACAGCGAGUCGCU---GU-UCA--UGCAACGUAAAGCACUGA .....((((...(((..((((((((((.......)))...(((.(((((.......))))---).)))..---..((((((...))))---))-)))--))))..)))..).))).. ( -35.00, z-score = 0.01, R) >consensus UCUUGAGUCGCCUCUUCUGCGUGAGUCGCCACAAGGCCACGCAAUCGGUGGCAGGCAGCG___AGUGCAG___C__CGACGCCUCCUUCCAGUUUCA_CUGCAGCGCAAGGCAGUCA ......((((((.....((((((....(((....)))))))))...))))))................................................................. (-16.24 = -15.99 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:40 2011