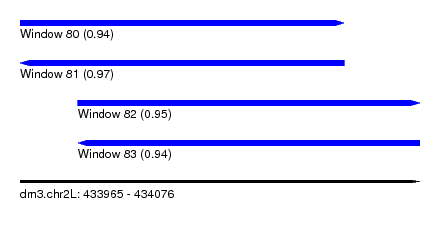
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 433,965 – 434,076 |
| Length | 111 |
| Max. P | 0.965623 |

| Location | 433,965 – 434,055 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 93.31 |
| Shannon entropy | 0.12594 |
| G+C content | 0.47424 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -27.28 |
| Energy contribution | -27.25 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 433965 90 + 23011544 UGUUUGGCCAUAACUGGCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUAUUUCU--CAGUCGCUUGUU ......((((....))))..((((((((((((((((.....))))))))(((....))).....((((..........--)))))))))))) ( -29.90, z-score = -2.54, R) >droSim1.chr2L 440426 90 + 22036055 UGUUUGGCCAUAACUGGCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUGUUUCU--CAGUCGCUUGUU ......((((....))))..((((((((((((((((.....))))))))(((....))).....((((..........--)))))))))))) ( -29.90, z-score = -2.13, R) >droSec1.super_14 417752 90 + 2068291 UGUUUGGCCAUAACUGGCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUGUUUUU--CAGUCGCUUGUU ......((((....))))..((((((((((((((((.....))))))))(((....))).....((((..........--)))))))))))) ( -29.90, z-score = -2.18, R) >droYak2.chr2L 418815 90 + 22324452 UGUUUAGCCAUAACUGGCACAGCAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUGUUUCU--CAGUCGCUUGUU ......((((....))))..((((((((((((((((.....))))))))(((....))).....((((..........--)))))))))))) ( -30.10, z-score = -2.13, R) >droEre2.scaffold_4929 486362 90 + 26641161 UGUUUGGCCAUAACUGGCACAGCAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUGUGCUGUUUUGUUUCU--CAGUCGCUUGUU ((...(((...(((.((((((((.....((((((((.....))))))))(((....))))))))))))))..)))...--)).......... ( -32.20, z-score = -2.11, R) >droAna3.scaffold_12943 224282 92 - 5039921 UGUUUGGCCAUAACUGGCACAACAAGUGGAAUUUCGGAAUUCGGAAUUCGGCAUUUGCCGCUGUUUGGUUUUGUUUCUGUCUGUCGCCUCUU .....(((.(((((.((((.(((.(((.((((((((.....))))))))(((....)))))).....))).))))...)).))).))).... ( -23.70, z-score = -0.51, R) >consensus UGUUUGGCCAUAACUGGCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUGUUUCU__CAGUCGCUUGUU ......((((....))))..((((((((((((((((.....))))))))(((....))).........................)))))))) (-27.28 = -27.25 + -0.03)

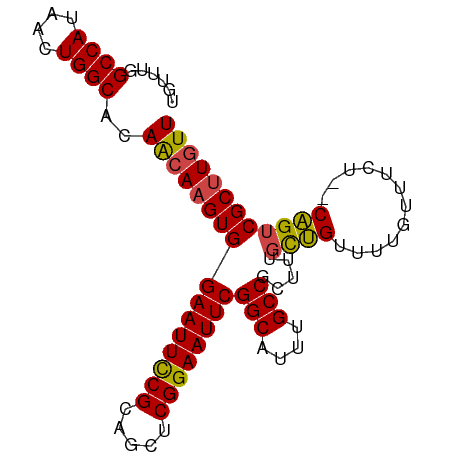
| Location | 433,965 – 434,055 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 93.31 |
| Shannon entropy | 0.12594 |
| G+C content | 0.47424 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -24.59 |
| Energy contribution | -24.73 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 433965 90 - 23011544 AACAAGCGACUG--AGAAAUAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGCCAGUUAUGGCCAAACA ............--.........(((((((...(((....)))((((((((.....))))))))...)))))))((((....))))...... ( -26.90, z-score = -2.14, R) >droSim1.chr2L 440426 90 - 22036055 AACAAGCGACUG--AGAAACAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGCCAGUUAUGGCCAAACA ............--.........(((((((...(((....)))((((((((.....))))))))...)))))))((((....))))...... ( -26.90, z-score = -1.95, R) >droSec1.super_14 417752 90 - 2068291 AACAAGCGACUG--AAAAACAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGCCAGUUAUGGCCAAACA ............--.........(((((((...(((....)))((((((((.....))))))))...)))))))((((....))))...... ( -26.90, z-score = -2.12, R) >droYak2.chr2L 418815 90 - 22324452 AACAAGCGACUG--AGAAACAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGCUGUGCCAGUUAUGGCUAAACA ............--.........(((((((...(((....)))((((((((.....))))))))...)))))))((((....))))...... ( -29.70, z-score = -2.91, R) >droEre2.scaffold_4929 486362 90 - 26641161 AACAAGCGACUG--AGAAACAAAACAGCACAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGCUGUGCCAGUUAUGGCCAAACA .....(((((((--.(........).((((((((((....)))((((((((.....)))))))).....))))))))))))...))...... ( -31.10, z-score = -3.24, R) >droAna3.scaffold_12943 224282 92 + 5039921 AAGAGGCGACAGACAGAAACAAAACCAAACAGCGGCAAAUGCCGAAUUCCGAAUUCCGAAAUUCCACUUGUUGUGCCAGUUAUGGCCAAACA .....(((((((.....................(((....)))(((((.((.....)).)))))...)))))))((((....))))...... ( -20.10, z-score = -1.07, R) >consensus AACAAGCGACUG__AGAAACAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGCCAGUUAUGGCCAAACA .......................((((((....(((....)))((((((((.....))))))))....))))))((((....))))...... (-24.59 = -24.73 + 0.14)

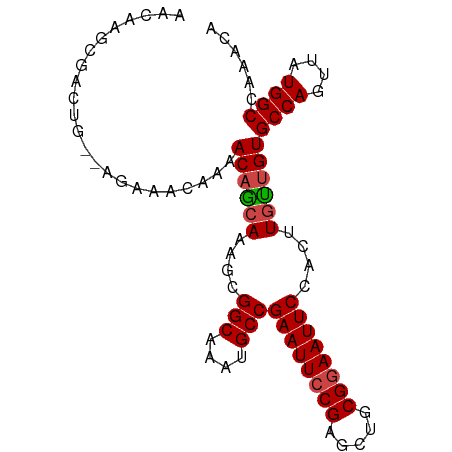
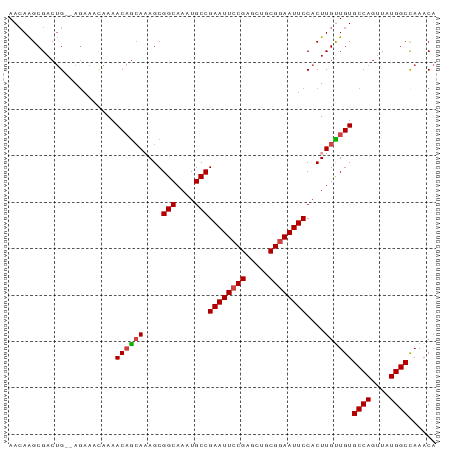
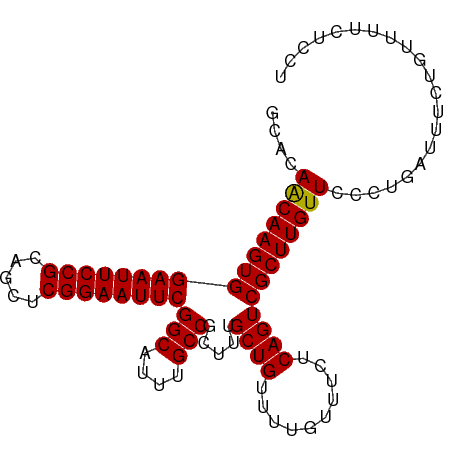
| Location | 433,981 – 434,076 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 97.89 |
| Shannon entropy | 0.03416 |
| G+C content | 0.47368 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -24.99 |
| Energy contribution | -24.80 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 433981 95 + 23011544 GCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUAUUUCUCAGUCGCUUGUUCCCUGAUUUCUGUUUUCUCCU ....((((((((((((((((.....))))))))(((....))).....((((..........))))))))))))..................... ( -24.80, z-score = -2.09, R) >droSim1.chr2L 440442 95 + 22036055 GCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUGUUUCUCAGUCGCUUGUUCCCUGAUUUCUGUUUUCUCCU ....((((((((((((((((.....))))))))(((....))).....((((..........))))))))))))..................... ( -24.80, z-score = -1.73, R) >droSec1.super_14 417768 95 + 2068291 GCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUGUUUUUCAGUCGCUUGUUCCCUGAUUUCUGUUUUCUCCU ....((((((((((((((((.....))))))))(((....))).....((((..........))))))))))))..................... ( -24.80, z-score = -1.78, R) >droEre2.scaffold_4929 486378 95 + 26641161 GCACAGCAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUGUGCUGUUUUGUUUCUCAGUCGCUUGUUCCCUGAUUUCUGUUUUCUCCU (((((((.....((((((((.....))))))))(((....))))))))))...........((((...........))))............... ( -30.00, z-score = -2.69, R) >consensus GCACAACAAGUGGAAUUCCGCAGCUCGGAAUUCGGCAUUUGCCGCUUUGCUGUUUUGUUUCUCAGUCGCUUGUUCCCUGAUUUCUGUUUUCUCCU ....((((((((((((((((.....))))))))(((....))).....((((..........))))))))))))..................... (-24.99 = -24.80 + -0.19)

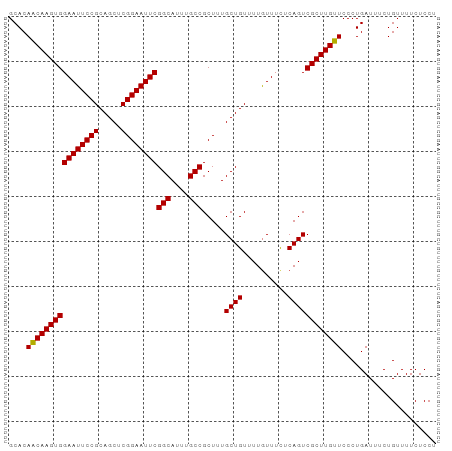
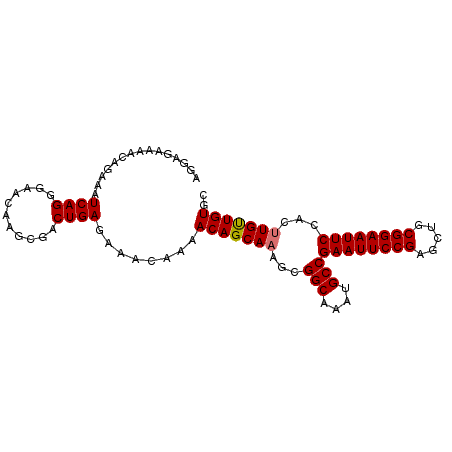
| Location | 433,981 – 434,076 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 97.89 |
| Shannon entropy | 0.03416 |
| G+C content | 0.47368 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -24.89 |
| Energy contribution | -24.95 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 433981 95 - 23011544 AGGAGAAAACAGAAAUCAGGGAACAAGCGACUGAGAAAUAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGC ...............((((...........))))........(((((((...(((....)))((((((((.....))))))))...))))))).. ( -24.80, z-score = -1.70, R) >droSim1.chr2L 440442 95 - 22036055 AGGAGAAAACAGAAAUCAGGGAACAAGCGACUGAGAAACAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGC ...............((((...........))))........(((((((...(((....)))((((((((.....))))))))...))))))).. ( -24.80, z-score = -1.53, R) >droSec1.super_14 417768 95 - 2068291 AGGAGAAAACAGAAAUCAGGGAACAAGCGACUGAAAAACAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGC ...............((((...........))))........(((((((...(((....)))((((((((.....))))))))...))))))).. ( -24.80, z-score = -1.71, R) >droEre2.scaffold_4929 486378 95 - 26641161 AGGAGAAAACAGAAAUCAGGGAACAAGCGACUGAGAAACAAAACAGCACAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGCUGUGC ...............((((...........))))...........((((((((((....)))((((((((.....)))))))).....))))))) ( -30.70, z-score = -3.20, R) >consensus AGGAGAAAACAGAAAUCAGGGAACAAGCGACUGAGAAACAAAACAGCAAAGCGGCAAAUGCCGAAUUCCGAGCUGCGGAAUUCCACUUGUUGUGC ...............((((...........))))........(((((((...(((....)))((((((((.....))))))))...))))))).. (-24.89 = -24.95 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:48 2011