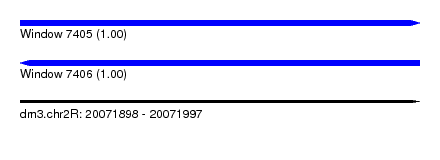
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,071,898 – 20,071,997 |
| Length | 99 |
| Max. P | 0.998946 |

| Location | 20,071,898 – 20,071,997 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 53.55 |
| Shannon entropy | 0.89433 |
| G+C content | 0.49747 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -16.08 |
| Energy contribution | -16.25 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.997368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
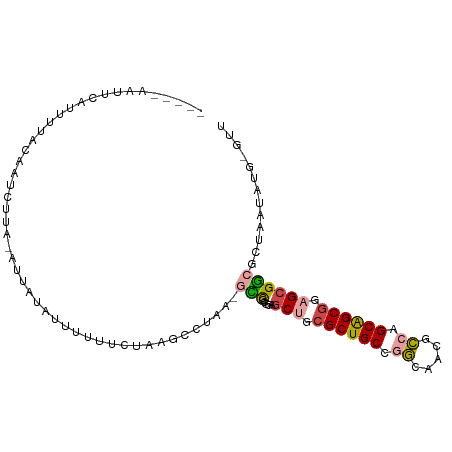
>dm3.chr2R 20071898 99 + 21146708 ---CUCUCUGAUUUUACAGUUUUUUAAUAUAUUUGGACUUAGGCUAA-UCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGCGUUCUUAUGUAUA ---....(((......))).......((((((..((((....((...-...))((.(((.((((((.((.....)).)))))).))).))))))..)))))). ( -31.70, z-score = -1.34, R) >droSim1.chr3h_random 590807 96 - 1452968 ----CAAU-CAUUUGGCGAUCUAA-AUACAAUGGUUUCUUAGCCUAA-GUCCAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUUUGCGUU ----....-......((.......-........((((((..((....-))..)))))).(((((((.(((...))).))))))).))(((((......))))) ( -32.70, z-score = -1.32, R) >droSec1.super_198 15265 97 - 31115 ----CAAUUCAUUUGGCGAUCUAA-AUACAAUGGUUUCUUAGCCUAA-GUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUUUGCGUU ----...........((.......-........((((((..((....-))..)))))).(((((((.(((...))).))))))).))(((((......))))) ( -35.50, z-score = -1.97, R) >droEre2.scaffold_4855 7530 86 + 242605 -----AUCUGGCGAUACAUUCUUA-AGUAUAUUUCGGCUAAACCUAAAACCGCGUAGCUGCGCUGCCAGCUACGCCAGCAGCGGAGCGGCUC----------- -----.....(((((((.......-.)))).....((.....))......)))((.(((.((((((..((...))..)))))).))).))..----------- ( -25.10, z-score = 0.06, R) >droAna3.scaffold_13230 2044919 86 - 3602488 -------------UUUCCAUUACAAUUUUUACUUAUUCUAGAACUUACGAUACAAGACUGCGCUGCUGCAGAGGGCAGCGGCUGAGCGUCAGCAAUAUA---- -------------..........................................(((.((((((((((.....))))))))...))))).........---- ( -19.00, z-score = -0.34, R) >droPer1.super_1295 3256 102 - 6930 UCUCUUCUAUAUUUUACAAUUGUG-GUUUGGGCUUUUGUAAGCCCAAAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGCGGCAGAACUAUGUGUU ..(((.((...........(((((-((((((((((....)))))).))))))))).(((.(((((((.(.....).))))))).))))).))).......... ( -43.30, z-score = -2.90, R) >consensus _____AAUUCAUUUUACAAUCUUA_AUUAUAUUUUUUCUAAGCCUAA_GCCGCGGAGCUGCGCUGCCGGCAACGCCAGCAGCGGAGCGGCGCUAAUAUG_GUU ................................................(((.....(((.((((((.((.....)).)))))).))))))............. (-16.08 = -16.25 + 0.17)

| Location | 20,071,898 – 20,071,997 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 53.55 |
| Shannon entropy | 0.89433 |
| G+C content | 0.49747 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -18.14 |
| Energy contribution | -17.95 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.998946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20071898 99 - 21146708 UAUACAUAAGAACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGA-UUAGCCUAAGUCCAAAUAUAUUAAAAAACUGUAAAAUCAGAGAG--- .............((.(((.(((((((((.....))))))))).)))...))(((-((......)))))...............(((......)))....--- ( -29.10, z-score = -2.30, R) >droSim1.chr3h_random 590807 96 + 1452968 AACGCAAAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUGGAC-UUAGGCUAAGAAACCAUUGUAU-UUAGAUCGCCAAAUG-AUUG---- .((((......)))).((.(((((((.(((...))).))))))).))........-...(((................-.......))).....-....---- ( -28.50, z-score = -0.07, R) >droSec1.super_198 15265 97 + 31115 AACGCAAAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGAC-UUAGGCUAAGAAACCAUUGUAU-UUAGAUCGCCAAAUGAAUUG---- .((((......)))).((.(((((((.(((...))).))))))).))........-...(((................-.......)))..........---- ( -28.50, z-score = -0.34, R) >droEre2.scaffold_4855 7530 86 - 242605 -----------GAGCCGCUCCGCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGUUUAGCCGAAAUAUACU-UAAGAAUGUAUCGCCAGAU----- -----------.(((......))).((((((((((((......)))))))(((.((((((((.((.......)).)))-))))).)))...)))))..----- ( -29.10, z-score = -0.33, R) >droAna3.scaffold_13230 2044919 86 + 3602488 ----UAUAUUGCUGACGCUCAGCCGCUGCCCUCUGCAGCAGCGCAGUCUUGUAUCGUAAGUUCUAGAAUAAGUAAAAAUUGUAAUGGAAA------------- ----((((..((((.((((.....(((((.....)))))))))))))..)))).......(((((..((((.......))))..))))).------------- ( -22.20, z-score = -1.17, R) >droPer1.super_1295 3256 102 + 6930 AACACAUAGUUCUGCCGCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUUUGGGCUUACAAAAGCCCAAAC-CACAAUUGUAAAAUAUAGAAGAGA .........(((((..(((.(((((((((.....))))))))).)))...(.((.(((((((((....))))))))))-).)............))))).... ( -40.10, z-score = -3.56, R) >consensus AAC_CAUAUAAGCGCCGCUCCGCUGCUGGCGUAGCCGGCAGCGCAGCUCCGCGGC_UUAGGCUAAGAAAAAAUAAAAU_UAAAAUUGUAAAAUCAAAU_____ .............((((((.(((((((((.....))))))))).))).....)))................................................ (-18.14 = -17.95 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:34 2011