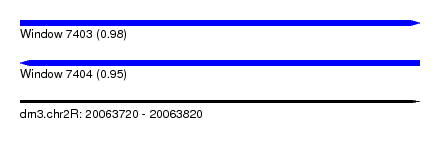
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,063,720 – 20,063,820 |
| Length | 100 |
| Max. P | 0.975667 |

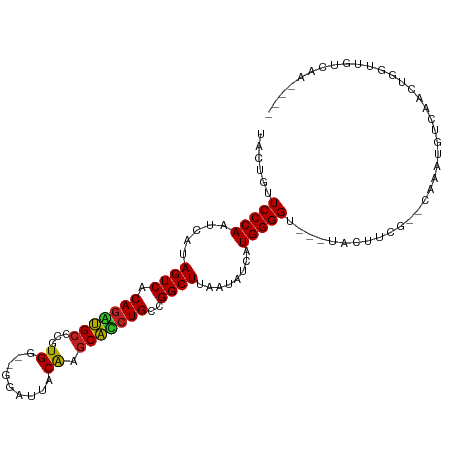
| Location | 20,063,720 – 20,063,820 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 68.00 |
| Shannon entropy | 0.69282 |
| G+C content | 0.47558 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -17.30 |
| Energy contribution | -16.07 |
| Covariance contribution | -1.23 |
| Combinations/Pair | 1.56 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20063720 100 + 21146708 ----UUGACAUCCAGUUGCCGUUUG--CGAAGUAGU-ACCCCAUGAUAUUAAGCCGGCAGGUGCCUGCAAUCC--CCACGGGCACCUGUGACUAUGAUUGGGAACAGCA ----............(((.((((.--(((.(((((-......((.........))(((((((((((......--...))))))))))).)))))..))).)))).))) ( -30.30, z-score = -0.65, R) >droGri2.scaffold_15245 680314 98 + 18325388 -UUGCUAGGAACAAGGUG----UUC--UUACUUA----CCCCAUGAUAUUAAGCCGGCAGACGCUUGUAAUUUAACCAGCGGCGUCUGUGACUAUGACUGGGAACAGUA -....(((((((.....)----)))--)))....----..................(((((((((.((..........))))))))))).......((((....)))). ( -25.00, z-score = -0.31, R) >droMoj3.scaffold_6496 25850488 101 + 26866924 ----UGCACAGUAUUAUGAGAUUUG--UAAGCUAU--ACCCCAAGAUAUUAAGCCGGCAGUCGUUCGUAUUUUAACCAGCGGCGACUGUGACUAUGACUGGGAACAGAA ----..((((((...........((--(..(((..--..............)))..)))((((((.((......)).)))))).)))))).......(((....))).. ( -22.89, z-score = -0.36, R) >droVir3.scaffold_12875 7136766 99 + 20611582 -----UGACUAUACUUC-AGAUUUUCAAAUCGU----ACCCCAUGAUAUUAAGCCGGCAGACGCAUGUAUUUUAACCAGCUGCGUCUGUGACUAUGACUGGGAACAGUA -----............-..........(((((----.....))))).........(((((((((.((..........))))))))))).......((((....)))). ( -21.40, z-score = -0.54, R) >droWil1.scaffold_180701 3592187 109 - 3904529 UUAUUGGAGAACCUUUUUUUUAUUGGUCCAUUUCUUUACCCCAUGAUAUUAAGCCGGCAGUUGCUCGAAAUUUAACCAAGAGCAGCUGUGACUAUGACUGGGAACAUUA ....(((((((((...........))).....)))))).((((.(.(((..((.((.(((((((((.............))))))))))).))))).)))))....... ( -23.52, z-score = -0.92, R) >droPer1.super_34 366772 105 - 916997 --GUUCAACAGUGUUUCUACACUUUCGAUUCCCCAUGACCCCAUGAUAUUAAGCCGGCAGACGCCCGUAUCCC--CCAUAAGCGUCUGUGACUUUGAUUGGGAACAGUA --..((...(((((....)))))...))(((((((((....))))..((((((...((((((((...(((...--..))).))))))))...)))))).)))))..... ( -25.80, z-score = -1.89, R) >dp4.chr3 1177196 105 - 19779522 --GUUCAACAGUGUUUCUACACUUUUGAUUCCCCAUGACCCCAUGAUAUUAAGCCGGCAGACGCCCGUAUACC--CCAUAAGCGUCUGUGACUUUGAUUGGGAACAGUA --..((((.(((((....))))).))))(((((((((....))))..((((((...((((((((...(((...--..))).))))))))...)))))).)))))..... ( -30.00, z-score = -3.36, R) >droAna3.scaffold_13266 5549230 90 - 19884421 ---------------GAAACAUUUUCAACAAAAC-UUACCCCAUGAUAUUAAGCCGGCAGAUGCUCGAAUCUC---CACGCGCAUUUGUGACUAUGAUUGGGAACAGUA ---------------.................((-(...((((....((((((.((.(((((((.((......---..)).))))))))).)).))))))))...))). ( -17.00, z-score = -0.63, R) >droEre2.scaffold_4845 21433758 97 + 22589142 ----UUGGCACUCAGUUGCCGUUUA--CGAAGUA----CCCCAUGAUAUUAAGCCGGCAGGUGCUUGCAAUCC--CCACGGGCACCUGUGACUAUGAUUGGGAACAACA ----..((((......)))).....--....((.----(((.((.(((........(((((((((((......--...)))))))))))...))).)).))).)).... ( -27.60, z-score = -0.23, R) >droYak2.chr2R 20035295 94 + 21139217 ----UUGGGAACUGGGUU---UAUA--CGAACUA----CCCCAUGAUAUUAAGCCGGCAGGUGCUUGCAAUCC--CCACGGGCACCUGUGACUAUGAUUGGGAACAACA ----.((((.....((((---(...--.))))).----.)))).............(((((((((((......--...)))))))))))........(((....))).. ( -25.50, z-score = -0.15, R) >droSec1.super_5 310899 97 + 5866729 ----UUGGCAUCCAGUUGCCGUUUG--CGAAGUA----CCCCAUGAUAUUAAGCCGGCAGGUGCUUGCAAUCC--CCACGGGCACCUGUGACUAUGAUUGGGAACAGCA ----..((((......))))...((--(...((.----(((.((.(((........(((((((((((......--...)))))))))))...))).)).))).)).))) ( -30.20, z-score = -0.21, R) >droSim1.chr2R 18553095 97 + 19596830 ----UUGGCAUCCAGUCGCCGUUUG--CGAAGUA----CCCCAUGAUAUUAAGCCGGCAGGUGCUUGCAAUCC--CCACGGGCACCUGUGACUAUGAUUGGGAACAGCA ----...((.(((((((((((...(--(..((((----........))))..))))))(((((((((......--...)))))))))........)))))))....)). ( -29.90, z-score = -0.08, R) >consensus ____UUGACAACCAGUUGACAUUUG__CGAAGUA___ACCCCAUGAUAUUAAGCCGGCAGAUGCUUGCAAUCC__CCACGGGCACCUGUGACUAUGAUUGGGAACAGUA .......................................((((........((.((.((((((((...............)))))))))).)).....))))....... (-17.30 = -16.07 + -1.23)

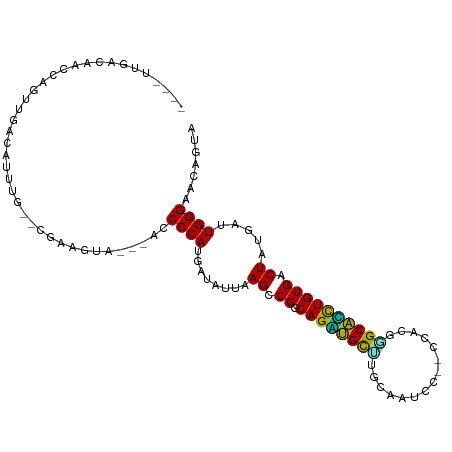
| Location | 20,063,720 – 20,063,820 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 68.00 |
| Shannon entropy | 0.69282 |
| G+C content | 0.47558 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -18.81 |
| Energy contribution | -17.84 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.55 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 20063720 100 - 21146708 UGCUGUUCCCAAUCAUAGUCACAGGUGCCCGUGG--GGAUUGCAGGCACCUGCCGGCUUAAUAUCAUGGGGU-ACUACUUCG--CAAACGGCAACUGGAUGUCAA---- (((((((((((.....((((.((((((((.(..(--...)..).))))))))..))))........))))((-........)--).)))))))............---- ( -31.92, z-score = -0.26, R) >droGri2.scaffold_15245 680314 98 - 18325388 UACUGUUCCCAGUCAUAGUCACAGACGCCGCUGGUUAAAUUACAAGCGUCUGCCGGCUUAAUAUCAUGGGG----UAAGUAA--GAA----CACCUUGUUCCUAGCAA- ((((..(((((.....((((.(((((((.....((......))..)))))))..))))........)))))----..)))).--(((----(.....)))).......- ( -26.52, z-score = -1.41, R) >droMoj3.scaffold_6496 25850488 101 - 26866924 UUCUGUUCCCAGUCAUAGUCACAGUCGCCGCUGGUUAAAAUACGAACGACUGCCGGCUUAAUAUCUUGGGGU--AUAGCUUA--CAAAUCUCAUAAUACUGUGCA---- .......(((((....((((.((((((....(.((......)).).))))))..)))).......)))))((--((((....--..............)))))).---- ( -19.77, z-score = 0.39, R) >droVir3.scaffold_12875 7136766 99 - 20611582 UACUGUUCCCAGUCAUAGUCACAGACGCAGCUGGUUAAAAUACAUGCGUCUGCCGGCUUAAUAUCAUGGGGU----ACGAUUUGAAAAUCU-GAAGUAUAGUCA----- (((((((((((.....((((.((((((((....((......)).))))))))..))))........))))).----))((((....)))).-..))))......----- ( -26.82, z-score = -1.63, R) >droWil1.scaffold_180701 3592187 109 + 3904529 UAAUGUUCCCAGUCAUAGUCACAGCUGCUCUUGGUUAAAUUUCGAGCAACUGCCGGCUUAAUAUCAUGGGGUAAAGAAAUGGACCAAUAAAAAAAAGGUUCUCCAAUAA ......(((((.....((((.(((.(((((.............))))).)))..))))........))))).........(((((...........)))))........ ( -23.14, z-score = -0.74, R) >droPer1.super_34 366772 105 + 916997 UACUGUUCCCAAUCAAAGUCACAGACGCUUAUGG--GGGAUACGGGCGUCUGCCGGCUUAAUAUCAUGGGGUCAUGGGGAAUCGAAAGUGUAGAAACACUGUUGAAC-- .....(((((.....(((((.((((((((..((.--....))..))))))))..))))).....((((....)))))))))((((.(((((....))))).))))..-- ( -36.20, z-score = -3.02, R) >dp4.chr3 1177196 105 + 19779522 UACUGUUCCCAAUCAAAGUCACAGACGCUUAUGG--GGUAUACGGGCGUCUGCCGGCUUAAUAUCAUGGGGUCAUGGGGAAUCAAAAGUGUAGAAACACUGUUGAAC-- .....(((((.....(((((.((((((((..((.--....))..))))))))..))))).....((((....)))))))))((((.(((((....))))).))))..-- ( -36.50, z-score = -3.53, R) >droAna3.scaffold_13266 5549230 90 + 19884421 UACUGUUCCCAAUCAUAGUCACAAAUGCGCGUG---GAGAUUCGAGCAUCUGCCGGCUUAAUAUCAUGGGGUAA-GUUUUGUUGAAAAUGUUUC--------------- .(((..(((((.....((((.((.((((.((..---......)).)))).))..))))........)))))..)-)).................--------------- ( -16.72, z-score = 0.89, R) >droEre2.scaffold_4845 21433758 97 - 22589142 UGUUGUUCCCAAUCAUAGUCACAGGUGCCCGUGG--GGAUUGCAAGCACCUGCCGGCUUAAUAUCAUGGGG----UACUUCG--UAAACGGCAACUGAGUGCCAA---- ........(((.....((((.(((((((..(..(--...)..)..)))))))..))))........)))((----(((((.(--(........)).)))))))..---- ( -27.92, z-score = 0.45, R) >droYak2.chr2R 20035295 94 - 21139217 UGUUGUUCCCAAUCAUAGUCACAGGUGCCCGUGG--GGAUUGCAAGCACCUGCCGGCUUAAUAUCAUGGGG----UAGUUCG--UAUA---AACCCAGUUCCCAA---- ......(((((.....((((.(((((((..(..(--...)..)..)))))))..))))........)))))----.......--....---..............---- ( -21.32, z-score = 1.28, R) >droSec1.super_5 310899 97 - 5866729 UGCUGUUCCCAAUCAUAGUCACAGGUGCCCGUGG--GGAUUGCAAGCACCUGCCGGCUUAAUAUCAUGGGG----UACUUCG--CAAACGGCAACUGGAUGCCAA---- (((.(((((((.....((((.(((((((..(..(--...)..)..)))))))..))))........)))))----.))...)--))...((((......))))..---- ( -30.22, z-score = 0.21, R) >droSim1.chr2R 18553095 97 - 19596830 UGCUGUUCCCAAUCAUAGUCACAGGUGCCCGUGG--GGAUUGCAAGCACCUGCCGGCUUAAUAUCAUGGGG----UACUUCG--CAAACGGCGACUGGAUGCCAA---- (((.(((((((.....((((.(((((((..(..(--...)..)..)))))))..))))........)))))----.))...)--))...((((......))))..---- ( -29.02, z-score = 0.74, R) >consensus UACUGUUCCCAAUCAUAGUCACAGAUGCCCGUGG__GGAUUACAAGCACCUGCCGGCUUAAUAUCAUGGGGU___UACUUCG__CAAAUGUCAACUGGUUGUCAA____ ......(((((.....((((.(((((((...((.........)).)))))))..))))........)))))...................................... (-18.81 = -17.84 + -0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:33 2011