| Sequence ID | dm3.chr2R |
|---|---|
| Location | 20,028,578 – 20,028,744 |
| Length | 166 |
| Max. P | 0.875185 |

| Location | 20,028,578 – 20,028,744 |
|---|---|
| Length | 166 |
| Sequences | 5 |
| Columns | 174 |
| Reading direction | forward |
| Mean pairwise identity | 66.84 |
| Shannon entropy | 0.59501 |
| G+C content | 0.47414 |
| Mean single sequence MFE | -46.24 |
| Consensus MFE | -30.06 |
| Energy contribution | -29.94 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
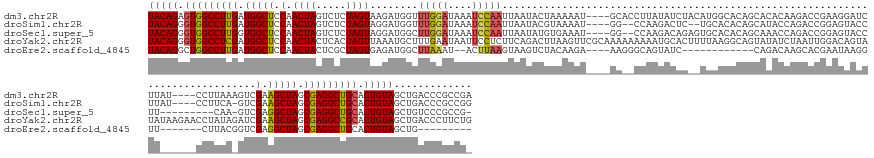
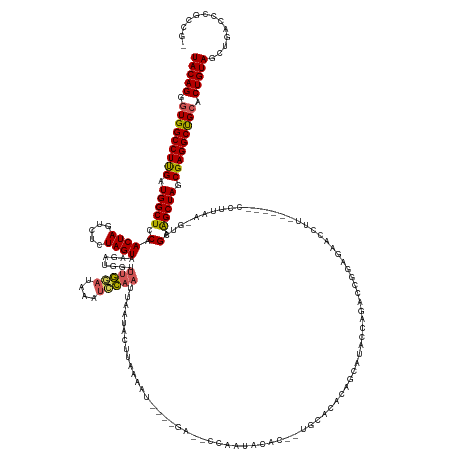
>dm3.chr2R 20028578 166 + 21146708 UACAGAGUGGCCUUGAUGGCUCCAACUAGUCUCUAGUAAGAUGGUUUGGAUAAAUCCAAUUAAUACUAAAAAU----GCACCUUAUAUCUACAUGGCACAGCACACAAGACCGAAGGAUCUUAU----CCUUAAAGUCGAAGCUAGCGAGGCUGCACUGUAGCUGACCCGCCGA (((((.(..((((((.(((((.(.(((((...)))))..(((..(..((((((((((...............(----(..((............))..))...............)))).))))----))..)..)))).))))).))))))..).)))))((......))... ( -44.41, z-score = -1.14, R) >droSim1.chr2R 18545148 161 + 19596830 UACAGGGUGGCCUUGAUGGCUCCAACUAGUCUCUAGUAGGAUGGUUUGGAUAAAUCCAAUUAAUACGUAAAAU----GG--CCAAGACUC--UGCACACAGCAUACCAGACCGGAGUACCUUAU----CCUUCA-GUCGAAGCUAGCGAGGCUGCACUGUAGCUGACCCGCCGG (((((.(..((((((.(((((((.(((((...))))).))).(((((((......(((.((........)).)----))--.........--(((.....)))..)))))))(((........)----))....-......)))).))))))..).)))))((......))... ( -48.30, z-score = -0.41, R) >droSec1.super_5 283850 157 + 5866729 UACAGGGUGGCCUUGGUGGCUCCAACUAGUCUCUAGUAGGAUGGCUUGGAUAAAUCCAAUUAAUAUGUGAAAU----GG--CCAAGACAGAGUGCACACAGCAAACCAGACCGGAGUACCUU---------CAA-GUCGAGGCUAGCGAGGCUGCACUGUAGCUGUCCCGCCG- (((((.(..((((((.(((((((.(((((...))))).)))(((((((((....(((...............(----(.--......))...(((.....))).........))).....))---------)))-))))..)))).))))))..).)))))((......))..- ( -49.70, z-score = -0.02, R) >droYak2.chr2R 20007714 174 + 21139217 UACAGGGUGGCCUCGAUGGCUCCAACUACUCACUAGUUAAAUGCUUUGAAUAAUUCCUCUUCAGACUUAAGUUCGCAAAAAAAAAUGCACUUUUAAGGCAGUAUAUCUAAUUGGACAGUAUAUAAGAACCUAUAGAUCGAAGCUAGCGAGGCCGCACUGUAGCUGACCCUUCUG (((((.(((((((((.(((((.(...((((..(((((((..(((((((((.............(((....))).(((........)))...))))))))).......)))))))..))))((((......))))....).))))).))))))))).)))))............. ( -46.50, z-score = -1.88, R) >droEre2.scaffold_4845 21406773 140 + 22589142 UACAGGCUGGCCUUGAUGGCUCCAACUACUCGCUAGUGAGAUGGCUUAAAU--ACUUAAGUAAGUCUACAAGA----AAGGGCAGUAUC------------CAGACAAGCACGAAUAAGGUU-------CUUACGGUCGAGGCUAGCGAGGCUGCACUGUAGCUG--------- (((((((.(((((((.((((..(.(((.(((.((.((.((((.((((((..--..))))))..)))))).)).----..))).)))...------------..(((......((((...)))-------).....))))..)))).))))))))).)))))....--------- ( -42.30, z-score = -0.30, R) >consensus UACAGGGUGGCCUUGAUGGCUCCAACUAGUCUCUAGUAAGAUGGUUUGGAUAAAUCCAAUUAAUACUUAAAAU____GA__CCAAUACAC__UGCACACAGCAUACCAGACCGGAGAACCUU______CCUUAA_GUCGAAGCUAGCGAGGCUGCACUGUAGCUGACCCGCCG_ (((((.(((((((((.(((((.(.((((.....))))........(((((....)))))...............................................................................).))))).))))))))).)))))............. (-30.06 = -29.94 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:29 2011