
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,799,370 – 3,799,462 |
| Length | 92 |
| Max. P | 0.588810 |

| Location | 3,799,370 – 3,799,462 |
|---|---|
| Length | 92 |
| Sequences | 13 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 63.92 |
| Shannon entropy | 0.83647 |
| G+C content | 0.42852 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -8.14 |
| Energy contribution | -8.35 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.28 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.588810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
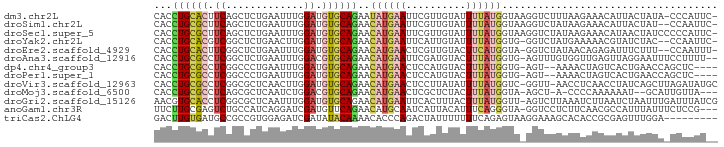
>dm3.chr2L 3799370 92 + 23011544 CACCUGCACUUCAGCUCUGAAUUUGGAUGUGCAGAAUAUGAAUUCGUUGUAUUUUAUGGUAAGGUCUUUAAGAAACAUUACUAUA-CCCAUUC- ...(((((((((((........))))).))))))((((..(.....)..)))).((((((((..((.....))....))))))))-.......- ( -18.60, z-score = -0.93, R) >droSim1.chr2L 3755398 91 + 22036055 CACCUGCGCUUCAGCUCUGAAUUUGGAUGUGCAGAACAUGAAUUCGUUGUAUUUUAUGGUAAGGUCUAUAAGAAACAUUACUAU--CCAAUUC- ...(((((((((((........))))).)))))).............(((.((((((((......)))))))).))).......--.......- ( -18.60, z-score = -0.73, R) >droSec1.super_5 1901453 93 + 5866729 CACCUGCGCUUCAGCUCUGAAUUUGGAUGUGCAGAACAUGAAUUCGUUGUAUUUUAUGGUAAGGUCUAUAAGAAACAUAACUAUCCCCCAUUC- ...(((((((((((........))))).)))))).............(((.((((((((......)))))))).)))................- ( -18.60, z-score = -0.59, R) >droYak2.chr2L 3801072 90 + 22324452 CACCUGCACGUCGGCUCUGAACUUGGAUGUGCAGAACAUGAAUUCAUUGUAUUUUAUGGUG-GGUCUAUGAAAAACGUAUCUAC--CCAAUUC- ...(((((((((.............)))))))))..((((((..........)))))).((-(((.((((.....))))...))--)))....- ( -23.02, z-score = -1.61, R) >droEre2.scaffold_4929 3844846 90 + 26641161 CACCUGCACUUCGGCUCUGAAUUUGGAUGUGCAGAACAUGAACUCGUUGUACUUCAUGGUA-GGUCUAUAACAGAGAUUUCUUU--CCAAUUU- ...(((((((((((........))))).))))))........(((((((((..((......-))..)))))).)))........--.......- ( -18.60, z-score = -0.08, R) >droAna3.scaffold_12916 1805039 91 + 16180835 CACCUGCGCCUCGGCUCUGAAUUUGGACGUGCAGAACAUGAAUUCGAUGUACUUUAUGGUG-AGUUUGUGGUUGAGUUAGGAAUUUCCUUUU-- ..((((...(((((((..(((((((..((((.((.((((.......)))).)).)))).))-)))))..))))))).))))...........-- ( -20.60, z-score = 0.06, R) >dp4.chr4_group3 6765826 87 - 11692001 CACCUGCGCCUCGGCCCUGAAUUUGGAUGUGCAGAACAUGAACUCCAUGUACUUUAUGGUG-AGU--AAAACUAGUCACUGAACCAGCUC---- ...((((((.((.............)).)))))).(((((.....))))).......((((-(.(--(....)).)))))..........---- ( -18.72, z-score = -0.11, R) >droPer1.super_1 3866044 87 - 10282868 CACCUGCGCCUCGGCCCUGAAUUUGGAUGUGCAGAACAUGAACUCCAUGUACUUUAUGGUG-AGU--AAAACUAGUCACUGAACCAGCUC---- ...((((((.((.............)).)))))).(((((.....))))).......((((-(.(--(....)).)))))..........---- ( -18.72, z-score = -0.11, R) >droVir3.scaffold_12963 5299161 92 - 20206255 CACCUGCGCCUCGGCGCUCAACUUGGAUGUGCAGAACAUGAACUCCUUAUAUUUUAUGGUC-GGUU-AACCUCAACCUAUCAGCUUAGAUAUGC .....((((....)))).......(((.((.((.....)).))))).(((((((..((((.-((((-......)))).))))....))))))). ( -20.80, z-score = -0.80, R) >droMoj3.scaffold_6500 14491261 86 - 32352404 CACCUGCGCCUCAGCGCUCAAUCUGGACGUGCAGAACAUGAACUCGCUCUACUUUAUGGUA-AGCU-A-CCCCAAAAAAU--GCAUUGUUA--- .....((((....))))........((((((((............(((.((((....))))-))).-.-..........)--))).)))).--- ( -16.85, z-score = -0.20, R) >droGri2.scaffold_15126 4470428 93 + 8399593 AACGUGCACCUCGGCGCUCAAUUUGGAUGUGCAGAACAUGAAUUCACUUUACUUUAUGGUU-AGUCUUAAAUCUUAAUCUAAUUUGAUUUAUCG ...((((......))))((((.((((((.....(((......)))(((..(((....))).-)))...........)))))).))))....... ( -13.50, z-score = 0.16, R) >anoGam1.chr3R 27765605 90 + 53272125 UUCUUGCGAGUCUGCCAUCAGGAUCGAUGUUCAGAACAUGCAAUCAUUACAUUUCAGGGUA-GGUCCUCUUCAACGCCAUUUAUUUCUCCG--- ...(((((..((((.((((......))))..))))...))))).............((((.-((......)).)).)).............--- ( -14.50, z-score = 0.55, R) >triCas2.ChLG4 2347829 85 + 13894384 GACUUGUGAUGCCGCCGUGGAGAUCGAUAUACAAAACACCCAGACUAUUUUUUUCAGAGUAAGGAAAGCACACCGCGAGUUUGGA--------- ((((((((.(((..(((((.................)))......(((((......))))).))...)))...))))))))....--------- ( -14.23, z-score = 0.73, R) >consensus CACCUGCGCCUCGGCUCUGAAUUUGGAUGUGCAGAACAUGAAUUCGUUGUAUUUUAUGGUA_GGUCUAUAACAAACAUAUCUAUU_CCUAUU__ ...((((((.((.............)).))))))..((((((..........)))))).................................... ( -8.14 = -8.35 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:42 2011