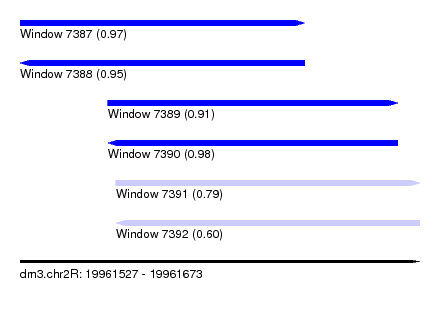
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,961,527 – 19,961,673 |
| Length | 146 |
| Max. P | 0.975074 |

| Location | 19,961,527 – 19,961,631 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.25 |
| Shannon entropy | 0.39604 |
| G+C content | 0.40447 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -18.83 |
| Energy contribution | -18.24 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
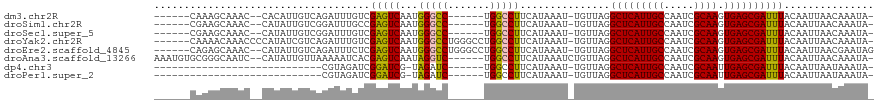
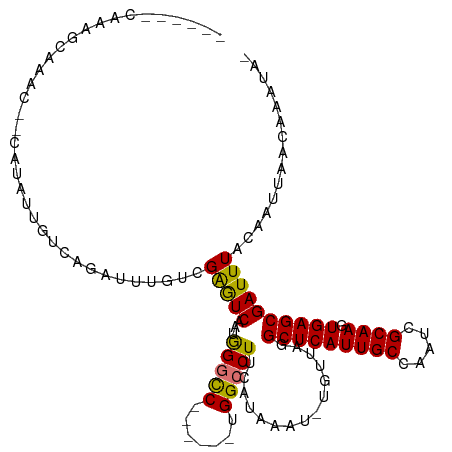
>dm3.chr2R 19961527 104 + 21146708 ------CAAAGCAAAC--CACAUUGUCAGAUUUGUCGAGUCAAUGGGCC------UGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA- ------....((((..--....))))...((((((..((.(((((((((------((((..........-.))))))))))))).)((((((......)))))).....)..)))))).- ( -29.10, z-score = -2.67, R) >droSim1.chr2R 18509050 104 + 19596830 ------CGAAGCAAAC--CAUAUUGUCGGAUUUGCCGAGUCAAUGGGCC------UGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA- ------....((((((--(........)).)))))...(.(((((((((------((((..........-.))))))))))))).)((((((......))))))...............- ( -30.40, z-score = -2.31, R) >droSec1.super_5 214770 104 + 5866729 ------CGAAGCAAAC--CAUAUUGUCGGAUUUGUCGAGUCAAUGGGCC------UGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA- ------((..((((..--....)))))).((((((..((.(((((((((------((((..........-.))))))))))))).)((((((......)))))).....)..)))))).- ( -29.60, z-score = -2.30, R) >droYak2.chr2R 19940373 112 + 21139217 ------CAAAACAAACCCCAUAUCGUCAGAUUUGUCGAGUCAAUGGGCCUGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA- ------..........(((((.(((.(......).)))....)))))..((((((((((..........-.))))))))))(((..((((((......))))))..)))..........- ( -29.50, z-score = -1.67, R) >droEre2.scaffold_4845 21336730 111 + 22589142 ------CAGAGCAAAC--CAUAUUGUCAGAUUUCUCGAGUCAAUGGGCCUGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACGAAUAG ------..(.((((.(--(.(((((.(.((....))..).)))))))..((((((((((..........-.)))))))))))))))((((((......))))))................ ( -27.30, z-score = -0.55, R) >droAna3.scaffold_13266 5444365 111 - 19884421 AAAUGUGCGGGCAAUC--CAUAUUGUUAAAAAUCACGAGUCAAUAGGUC------UGGCCUUCAUAAAUCUGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA- ...((((.((((...(--(.(((((...............)))))))..------..)))).))))....((((((((.....)))((((((......))))))......)))))....- ( -27.46, z-score = -2.10, R) >dp4.chr3 7652724 83 + 19779522 ----------------------------CGUAGAUCGGAUCG-UAGAUC------UGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAUUGAGCGAUUUACAAUUAAUAAAUA- ----------------------------.(.((..((((((.-..))))------))..)).)...(((-(((..(((.....)))((((((......)))))).))))))........- ( -22.40, z-score = -2.59, R) >droPer1.super_2 7852303 83 + 9036312 ----------------------------CGUAGAUCGGAUCG-UAGAUC------UGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAUUGAGCGAUUUACAAUUAAUAAAUA- ----------------------------.(.((..((((((.-..))))------))..)).)...(((-(((..(((.....)))((((((......)))))).))))))........- ( -22.40, z-score = -2.59, R) >consensus ______CAAAGCAAAC__CAUAUUGUCAGAUUUGUCGAGUCAAUGGGCC______UGGCCUUCAUAAAU_UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA_ ....................................(((((...(((((.......)))))...............(((((((((.....)))).))))))))))............... (-18.83 = -18.24 + -0.59)
| Location | 19,961,527 – 19,961,631 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Shannon entropy | 0.39604 |
| G+C content | 0.40447 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -16.56 |
| Energy contribution | -16.43 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19961527 104 - 21146708 -UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCA------GGCCCAUUGACUCGACAAAUCUGACAAUGUG--GUUUGCUUUG------ -...((((((......((((((......))))))(((.....))))))))-)......(((((.(------(((((((((..(((.......)))))))).)--))))))))).------ ( -28.20, z-score = -2.20, R) >droSim1.chr2R 18509050 104 - 19596830 -UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCA------GGCCCAUUGACUCGGCAAAUCCGACAAUAUG--GUUUGCUUCG------ -...((((((......((((((......))))))(((.....))))))))-).....((((..((------((((.((((..((((.....))))))))..)--))))))))).------ ( -28.70, z-score = -1.97, R) >droSec1.super_5 214770 104 - 5866729 -UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCA------GGCCCAUUGACUCGACAAAUCCGACAAUAUG--GUUUGCUUCG------ -...((((((......((((((......))))))(((.....))))))))-).....((((..((------((((.((((..(((.......)))))))..)--))))))))).------ ( -26.10, z-score = -1.78, R) >droYak2.chr2R 19940373 112 - 21139217 -UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCAGGCCCAUUGACUCGACAAAUCUGACGAUAUGGGGUUUGUUUUG------ -..((..(.((((((.((((((......))))))(((.....)))..)))-))).)..))((((.......)))).........(((((((((.(.....).)))))))))...------ ( -30.20, z-score = -1.27, R) >droEre2.scaffold_4845 21336730 111 - 22589142 CUAUUCGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCAGGCCCAUUGACUCGAGAAAUCUGACAAUAUG--GUUUGCUCUG------ ....((((.((((((.((((((......))))))(((.....)))..)))-))).))))(((((((((....))).((((..((((....)).)))))).))--))))......------ ( -30.80, z-score = -1.93, R) >droAna3.scaffold_13266 5444365 111 + 19884421 -UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACAGAUUUAUGAAGGCCA------GACCUAUUGACUCGUGAUUUUUAACAAUAUG--GAUUGCCCGCACAUUU -.((((((((......((((((......))))))(((.....))))))))))).......((.((------(.(((((((...............))))).)--).))).))........ ( -23.76, z-score = -1.13, R) >dp4.chr3 7652724 83 - 19779522 -UAUUUAUUAAUUGUAAAUCGCUCAAUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCA------GAUCUA-CGAUCCGAUCUACG---------------------------- -..(((((.((((((.((((((......))))))(((.....)))..)))-))).)))))....(------((((..-......)))))...---------------------------- ( -21.60, z-score = -3.17, R) >droPer1.super_2 7852303 83 - 9036312 -UAUUUAUUAAUUGUAAAUCGCUCAAUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCA------GAUCUA-CGAUCCGAUCUACG---------------------------- -..(((((.((((((.((((((......))))))(((.....)))..)))-))).)))))....(------((((..-......)))))...---------------------------- ( -21.60, z-score = -3.17, R) >consensus _UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA_AUUUAUGAAGGCCA______GGCCCAUUGACUCGACAAAUCUGACAAUAUG__GUUUGCUUUG______ ......(((((((((.((((((......)))))).)))))(.((((.............)))))..............))))...................................... (-16.56 = -16.43 + -0.13)
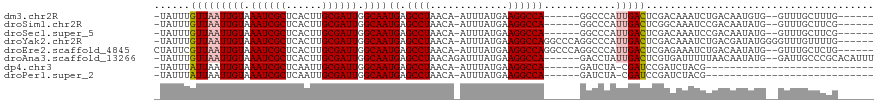
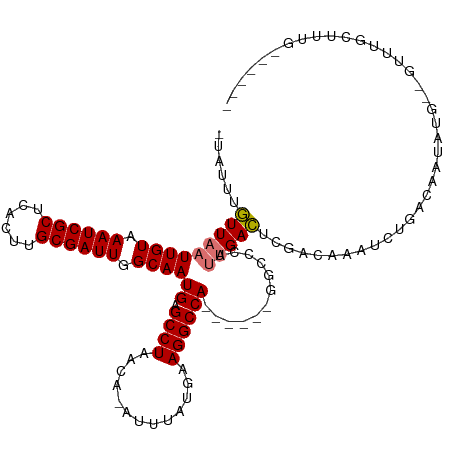
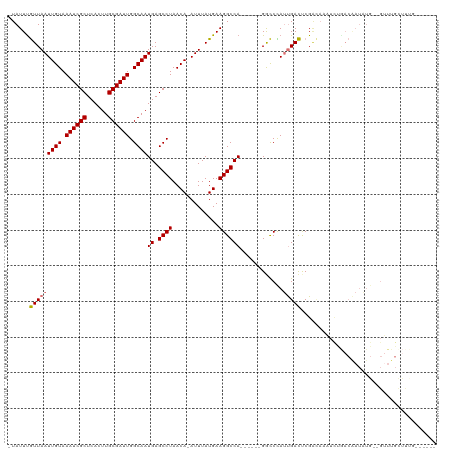
| Location | 19,961,559 – 19,961,665 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.05 |
| Shannon entropy | 0.30894 |
| G+C content | 0.36926 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -16.13 |
| Energy contribution | -16.74 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19961559 106 + 21146708 ------CAAUGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCCAGA-GCCCCUCUAUUUAUAGAUAUGUUUAUAC ------....((((((((........(((-(((..(((.....)))((((((......)))))).))))))........------.)))).-)))).......((((((....)))))). ( -27.93, z-score = -2.67, R) >droSim1.chr2R 18509082 106 + 19596830 ------CAAUGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCCAGA-GCCCCUCUAUUUAUAGAUAUGUUUAUAC ------....((((((((........(((-(((..(((.....)))((((((......)))))).))))))........------.)))).-)))).......((((((....)))))). ( -27.93, z-score = -2.67, R) >droSec1.super_5 214802 106 + 5866729 ------CAAUGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCCAGA-GCCCCUCUAUUUAUAGAUUUGUUUAUAC ------....((((((((........(((-(((..(((.....)))((((((......)))))).))))))........------.)))).-)))).......((((((....)))))). ( -27.93, z-score = -2.72, R) >droYak2.chr2R 19940407 112 + 21139217 CAAUGGGCCUGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCCAGU-GCCCCUCUAUUUAUAGAUAUGUUUAUAC ....(((((((((...(((((.((.....-))..))))).((((..((((((......))))))..)))).........------))))).-)))).......((((((....)))))). ( -32.90, z-score = -2.45, R) >droEre2.scaffold_4845 21336762 118 + 22589142 CAAUGGGCCUGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACGAAUAGCCAUGCCCAGA-GCCCCUCUAUUUUUAGAUAUGUUUAUAC ....((((((((((.((((.(((...(((-(((..(((.....)))((((((......)))))).))))))...)))..)))).)))))).-)))).((((....))))........... ( -44.60, z-score = -5.00, R) >droAna3.scaffold_13266 5444403 108 - 19884421 ------CAAUAGGUCUGGCCUUCAUAAAUCUGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCUAGAUACUGGUUUAUUAUUAGAUAUGUUUAUAC ------...(((((..(((((.((......))..))))).((((..((((((......))))))..))))........)------))))....(((((.....)))))............ ( -20.60, z-score = -0.56, R) >dp4.chr3 7652738 96 + 19779522 ---------UAGAUCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAUUGAGCGAUUUACAAUUAAUAAAUA--------CAUA------CUUAUUUAUAGAUUUUAUAUUGU ---------.(((((((((((.((.....-))..))))).((((..((((((......))))))..))))..(((((((--------....------..)))))))))))))........ ( -19.30, z-score = -1.93, R) >droPer1.super_2 7852317 96 + 9036312 ---------UAGAUCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAUUGAGCGAUUUACAAUUAAUAAAUA--------CAUA------CUUAUUUAUAGAUUUUAUAUUGU ---------.(((((((((((.((.....-))..))))).((((..((((((......))))))..))))..(((((((--------....------..)))))))))))))........ ( -19.30, z-score = -1.93, R) >consensus ______CAAUGGGCCUGGCCUUCAUAAAU_UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA______CCCAGA_GCCCCUCUAUUUAUAGAUAUGUUUAUAC ...........(((((((............((((((((.....)))((((((......))))))......)))))...........))))..)))......................... (-16.13 = -16.74 + 0.61)
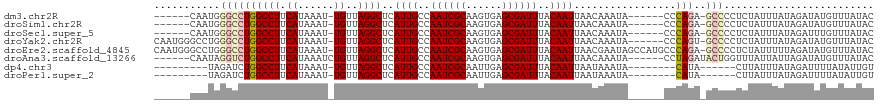
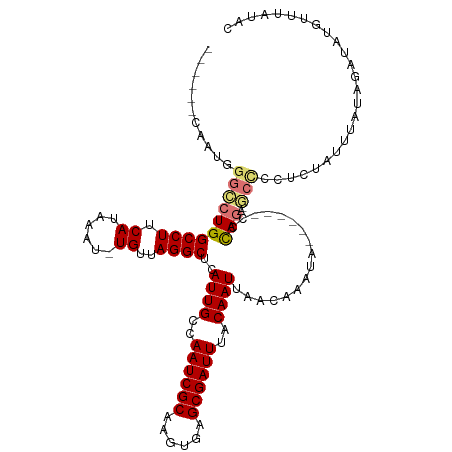
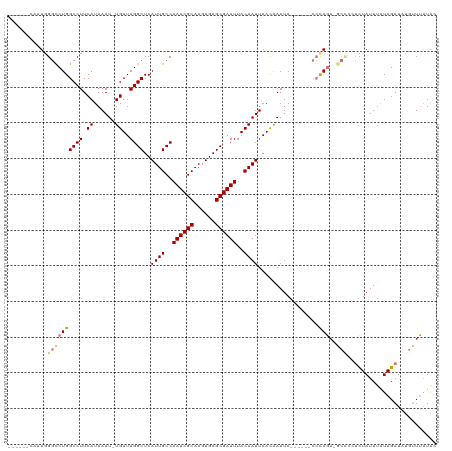
| Location | 19,961,559 – 19,961,665 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.05 |
| Shannon entropy | 0.30894 |
| G+C content | 0.36926 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -17.95 |
| Energy contribution | -18.11 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19961559 106 - 21146708 GUAUAAACAUAUCUAUAAAUAGAGGGGC-UCUGGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCAUUG------ ...........((((....)))).((((-.((((.------..((..(.((((((.((((((......))))))(((.....)))..)))-))).)..))..))))))))....------ ( -33.70, z-score = -3.19, R) >droSim1.chr2R 18509082 106 - 19596830 GUAUAAACAUAUCUAUAAAUAGAGGGGC-UCUGGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCAUUG------ ...........((((....)))).((((-.((((.------..((..(.((((((.((((((......))))))(((.....)))..)))-))).)..))..))))))))....------ ( -33.70, z-score = -3.19, R) >droSec1.super_5 214802 106 - 5866729 GUAUAAACAAAUCUAUAAAUAGAGGGGC-UCUGGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCAUUG------ ...........((((....)))).((((-.((((.------..((..(.((((((.((((((......))))))(((.....)))..)))-))).)..))..))))))))....------ ( -33.70, z-score = -3.31, R) >droYak2.chr2R 19940407 112 - 21139217 GUAUAAACAUAUCUAUAAAUAGAGGGGC-ACUGGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCAGGCCCAUUG ...........((((....)))).((((-.(((((------(........(((((.((((((......)))))).)))))(.((((....-........)))))..)))))))))).... ( -37.60, z-score = -3.13, R) >droEre2.scaffold_4845 21336762 118 - 22589142 GUAUAAACAUAUCUAAAAAUAGAGGGGC-UCUGGGCAUGGCUAUUCGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCAGGCCCAUUG ...........((((....)))).((((-.((((((.(((((.(((((.((((((.((((((......))))))(((.....)))..)))-))).)))))))))).)))))))))).... ( -51.60, z-score = -6.29, R) >droAna3.scaffold_13266 5444403 108 + 19884421 GUAUAAACAUAUCUAAUAAUAAACCAGUAUCUAGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACAGAUUUAUGAAGGCCAGACCUAUUG------ ...............................((((------(........(((((.((((((......)))))).)))))(.((((..((......)).)))))..)))))...------ ( -21.90, z-score = -1.26, R) >dp4.chr3 7652738 96 - 19779522 ACAAUAUAAAAUCUAUAAAUAAG------UAUG--------UAUUUAUUAAUUGUAAAUCGCUCAAUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGAUCUA--------- ..........((((...((((((------(...--------.))))))).(((((.((((((......)))))).)))))(.((((....-........)))))))))...--------- ( -18.60, z-score = -1.51, R) >droPer1.super_2 7852317 96 - 9036312 ACAAUAUAAAAUCUAUAAAUAAG------UAUG--------UAUUUAUUAAUUGUAAAUCGCUCAAUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGAUCUA--------- ..........((((...((((((------(...--------.))))))).(((((.((((((......)))))).)))))(.((((....-........)))))))))...--------- ( -18.60, z-score = -1.51, R) >consensus GUAUAAACAUAUCUAUAAAUAGAGGGGC_UCUGGG______UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA_AUUUAUGAAGGCCAGGCCCAUUG______ ...............................((((...............(((((.((((((......)))))).)))))(.((((.............)))))...))))......... (-17.95 = -18.11 + 0.16)
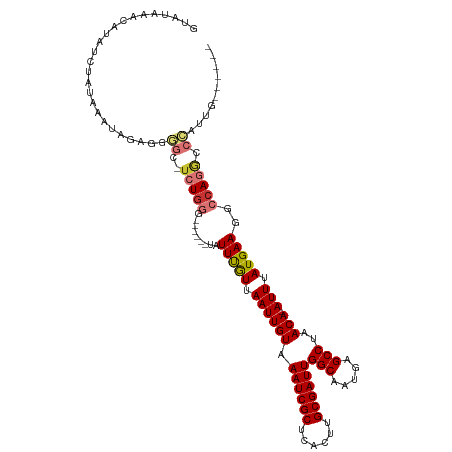
| Location | 19,961,562 – 19,961,673 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.87 |
| Shannon entropy | 0.26374 |
| G+C content | 0.37135 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -19.65 |
| Energy contribution | -20.96 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19961562 111 + 21146708 -UGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCCAGA-GCCCCUCUAUUUAUAGAUAUGUUUAUACAUGGGCAU -.((((((((........(((-(((..(((.....)))((((((......)))))).))))))........------.)))).-)))).(((((.((((((....)))))).)))))... ( -31.83, z-score = -2.21, R) >droSim1.chr2R 18509085 111 + 19596830 -UGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCCAGA-GCCCCUCUAUUUAUAGAUAUGUUUAUACAUGGGCAU -.((((((((........(((-(((..(((.....)))((((((......)))))).))))))........------.)))).-)))).(((((.((((((....)))))).)))))... ( -31.83, z-score = -2.21, R) >droSec1.super_5 214805 111 + 5866729 -UGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCCAGA-GCCCCUCUAUUUAUAGAUUUGUUUAUACAUGGGCAU -.((((((((........(((-(((..(((.....)))((((((......)))))).))))))........------.)))).-)))).(((((.((((((....)))))).)))))... ( -31.83, z-score = -2.27, R) >droYak2.chr2R 19940415 112 + 21139217 CUGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCCAGU-GCCCCUCUAUUUAUAGAUAUGUUUAUACAUGGGCAU (((((...(((((.((.....-))..))))).((((..((((((......))))))..)))).........------)))))(-((((.((((....)))).((((....))))))))). ( -32.70, z-score = -2.29, R) >droEre2.scaffold_4845 21336770 118 + 22589142 CUGGGCCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACGAAUAGCCAUGCCCAGA-GCCCCUCUAUUUUUAGAUAUGUUUAUACAUGGGCAU ((((((.((((.(((...(((-(((..(((.....)))((((((......)))))).))))))...)))..)))).)))))).-((((.((((....)))).((((....)))))))).. ( -44.50, z-score = -4.89, R) >droAna3.scaffold_13266 5444406 113 - 19884421 -UAGGUCUGGCCUUCAUAAAUCUGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA------CCUAGAUACUGGUUUAUUAUUAGAUAUGUUUAUACAUGGGCAU -........((((......(((((((((((.....)))((((((......))))))......))))....(------((........)))........))))((((....)))))))).. ( -23.50, z-score = -0.34, R) >dp4.chr3 7652738 104 + 19779522 -UAGAUCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAUUGAGCGAUUUACAAUUAAUAAAUA--------------CAUACUUAUUUAUAGAUUUUAUAUUGUAUAGGUAU -........((((..((((((-(((..(((.....)))((((((......)))))).)))))).(((((((--------------......)))))))...........)))..)))).. ( -20.90, z-score = -1.59, R) >droPer1.super_2 7852317 104 + 9036312 -UAGAUCUGGCCUUCAUAAAU-UGUUAGGCUCAUUGCCAAUCGCAAUUGAGCGAUUUACAAUUAAUAAAUA--------------CAUACUUAUUUAUAGAUUUUAUAUUGUAUAGGUAU -........((((..((((((-(((..(((.....)))((((((......)))))).)))))).(((((((--------------......)))))))...........)))..)))).. ( -20.90, z-score = -1.59, R) >consensus _UGGGCCUGGCCUUCAUAAAU_UGUUAGGCUCAUUGCCAAUCGCAAGUGAGCGAUUUACAAUUAACAAAUA______CCCAGA_GCCCCUCUAUUUAUAGAUAUGUUUAUACAUGGGCAU ..((((((((............((((((((.....)))((((((......))))))......)))))...........))))..)))).(((((.((((((....)))))).)))))... (-19.65 = -20.96 + 1.32)

| Location | 19,961,562 – 19,961,673 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.87 |
| Shannon entropy | 0.26374 |
| G+C content | 0.37135 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -16.64 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19961562 111 - 21146708 AUGCCCAUGUAUAAACAUAUCUAUAAAUAGAGGGGC-UCUGGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCA- ......((((....)))).((((....)))).((((-.((((.------..((..(.((((((.((((((......))))))(((.....)))..)))-))).)..))..)))))))).- ( -34.50, z-score = -2.17, R) >droSim1.chr2R 18509085 111 - 19596830 AUGCCCAUGUAUAAACAUAUCUAUAAAUAGAGGGGC-UCUGGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCA- ......((((....)))).((((....)))).((((-.((((.------..((..(.((((((.((((((......))))))(((.....)))..)))-))).)..))..)))))))).- ( -34.50, z-score = -2.17, R) >droSec1.super_5 214805 111 - 5866729 AUGCCCAUGUAUAAACAAAUCUAUAAAUAGAGGGGC-UCUGGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCA- ...................((((....)))).((((-.((((.------..((..(.((((((.((((((......))))))(((.....)))..)))-))).)..))..)))))))).- ( -33.70, z-score = -2.05, R) >droYak2.chr2R 19940415 112 - 21139217 AUGCCCAUGUAUAAACAUAUCUAUAAAUAGAGGGGC-ACUGGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCAG .(((((((((....)))).((((....)))).))))-)(((((------(........(((((.((((((......)))))).)))))(.((((....-........)))))..)))))) ( -37.20, z-score = -3.13, R) >droEre2.scaffold_4845 21336770 118 - 22589142 AUGCCCAUGUAUAAACAUAUCUAAAAAUAGAGGGGC-UCUGGGCAUGGCUAUUCGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGGCCCAG ..((((((((....)))).((((....)))).))))-.((((((.(((((.(((((.((((((.((((((......))))))(((.....)))..)))-))).)))))))))).)))))) ( -49.60, z-score = -5.84, R) >droAna3.scaffold_13266 5444406 113 + 19884421 AUGCCCAUGUAUAAACAUAUCUAAUAAUAAACCAGUAUCUAGG------UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACAGAUUUAUGAAGGCCAGACCUA- .((((.((((....)))).....(((((.(((.(((((....)------)))).))).))))).((((((......)))))))))).((.((((..((......)).))))))......- ( -24.90, z-score = -1.49, R) >dp4.chr3 7652738 104 - 19779522 AUACCUAUACAAUAUAAAAUCUAUAAAUAAGUAUG--------------UAUUUAUUAAUUGUAAAUCGCUCAAUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGAUCUA- ..................((((...(((((((...--------------.))))))).(((((.((((((......)))))).)))))(.((((....-........)))))))))...- ( -18.60, z-score = -1.26, R) >droPer1.super_2 7852317 104 - 9036312 AUACCUAUACAAUAUAAAAUCUAUAAAUAAGUAUG--------------UAUUUAUUAAUUGUAAAUCGCUCAAUUGCGAUUGGCAAUGAGCCUAACA-AUUUAUGAAGGCCAGAUCUA- ..................((((...(((((((...--------------.))))))).(((((.((((((......)))))).)))))(.((((....-........)))))))))...- ( -18.60, z-score = -1.26, R) >consensus AUGCCCAUGUAUAAACAUAUCUAUAAAUAGAGGGGC_UCUGGG______UAUUUGUUAAUUGUAAAUCGCUCACUUGCGAUUGGCAAUGAGCCUAACA_AUUUAUGAAGGCCAGGCCCA_ .......................................((((...............(((((.((((((......)))))).)))))(.((((.............)))))...)))). (-16.64 = -17.48 + 0.84)
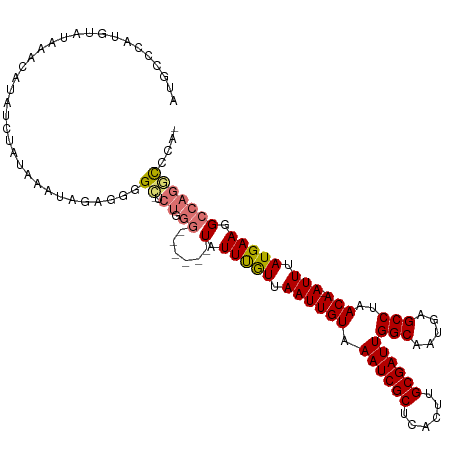
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:22 2011