
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,956,037 – 19,956,127 |
| Length | 90 |
| Max. P | 0.536730 |

| Location | 19,956,037 – 19,956,127 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 49.89 |
| Shannon entropy | 1.00301 |
| G+C content | 0.41323 |
| Mean single sequence MFE | -24.19 |
| Consensus MFE | 0.00 |
| Energy contribution | 0.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 0.00 |
| Mean z-score | -2.46 |
| Structure conservation index | -0.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19956037 90 - 21146708 ---------CUUUAGCUAUAGCU---AAAGGAAAUCGACAAAAGU-CGUAAUCGGCUUGCGGAAUACCC--CUGGCAUUCCGCAGGUGCAGCCACGAAGCCGAUU ---------(((((((....)))---)))).....((((....))-)).((((((((((((((((.((.--..)).))))))).((.....))...))))))))) ( -37.40, z-score = -4.82, R) >droSim1.chr2R 18503098 93 - 19596830 ---------CUUUAGCUAUAGCUAAAAAAAAAAAACGACAAAAAU-CGUUAUCGGUUUCCGGAAUACCC--CUGGCAUUCCGCAGGUGCAGCCACGAAGCCGAUU ---------.((((((....)))))).......(((((......)-))))(((((((((((((((.((.--..)).))))))..((.....))..))))))))). ( -29.20, z-score = -3.98, R) >droSec1.super_5 209339 90 - 5866729 ---------CUUUAGCUAUAGCU---AAAAGAAAUCGACAAAAAU-CGUAAUCGGUUUUCGGAAUACCC--CUGGCAUUCCGCAGGUGCAGCCACGAAGCCGAUU ---------.((((((....)))---)))......(((......)-)).((((((((((((((((.((.--..)).))))))..((.....))..)))))))))) ( -24.60, z-score = -2.02, R) >droYak2.chr2R 19934934 89 - 21139217 ----------CUUGGCAAAAGCU---AAGUAAUAUUGGCAAAAAU-CGUAAUCGGUUUGCGGAAUACCC--CUGACAUUCCGCAGGUGCAGCCACGAAGCCGAUU ----------((((((....)))---)))....((((((..((((-((....))))))(((((((....--.....))))))).((.....)).....)))))). ( -27.60, z-score = -2.20, R) >droEre2.scaffold_4845 21331348 83 - 22589142 ---------CUUUAGCUAAAGCUA--AAACAAAAUCGAUAAAAAU-CGUAAU--------GGAAUACCC--CUGACCUGCUGCAGGUGUAGCCACGAAGCCGAUU ---------.((((((....))))--))....(((((.......(-(((...--------((..(((.(--(((........)))).))).))))))...))))) ( -16.80, z-score = -1.67, R) >droAna3.scaffold_13266 5438748 86 + 19884421 ----------CUUUUAAAAAG-----AAACUAUUUUGUUAGGUAUGCUUAACAAAGAAAACGGAUAGGU--GU-AUACAGCACAUUCCAACGCAACAGGUCGAU- ----------..........(-----(.....((((((((((....)))))))))).....(((...((--((-.....))))..)))...........))...- ( -17.20, z-score = -1.96, R) >droVir3.scaffold_12875 12050517 94 - 20611582 UUGCAGUCAUAUCACACAAGAAUGUUAGAUAUGUU--GCAAAUGUUUUUAUCCAAUUGUGCAUAUAAA---AUAGUGUGUCGAAUAUAUCGGUUCGGAU------ ((((((.((((((..(((....)))..))))))))--))))........((((......((((((...---...)))))).((((......))))))))------ ( -21.60, z-score = -1.64, R) >droGri2.scaffold_15245 11921623 92 - 18325388 UUAUUGCCACUUCCAUGAGGGCUUAUAAACGUCGCAAAUCAAAAUUUAUUAAAAUUUUAAGGUGUGUUCCGCUAGUAUCGGAACAGUGAAAU------------- ((((.(((.(((....))))))..))))...((((.....(((((((....)))))))......(((((((.......)))))))))))...------------- ( -19.10, z-score = -1.40, R) >consensus _________CUUUAGCUAAAGCU___AAACAAAAUCGACAAAAAU_CGUAAUCGGUUUACGGAAUACCC__CUGGCAUUCCGCAGGUGCAGCCACGAAGCCGAUU ......................................................................................................... ( 0.00 = 0.00 + 0.00)

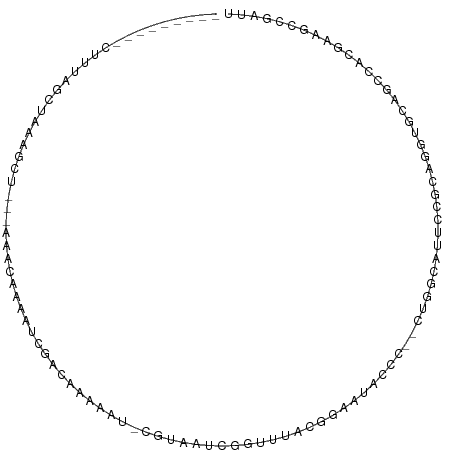
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:17 2011