| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,943,603 – 19,943,719 |
| Length | 116 |
| Max. P | 0.945569 |

| Location | 19,943,603 – 19,943,719 |
|---|---|
| Length | 116 |
| Sequences | 7 |
| Columns | 140 |
| Reading direction | reverse |
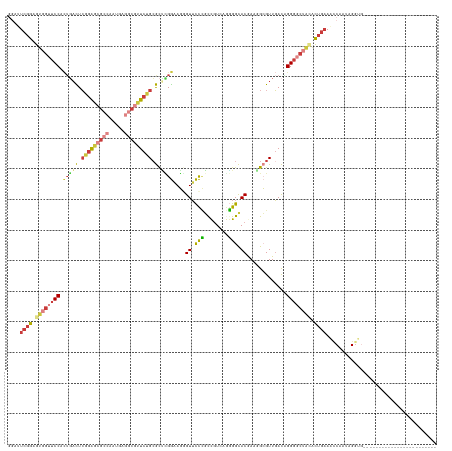
| Mean pairwise identity | 66.30 |
| Shannon entropy | 0.60164 |
| G+C content | 0.57230 |
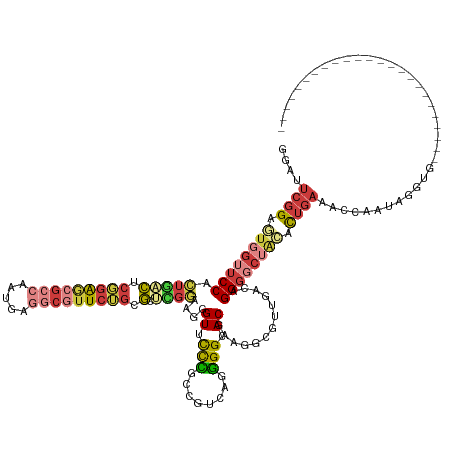
| Mean single sequence MFE | -45.81 |
| Consensus MFE | -23.95 |
| Energy contribution | -25.27 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
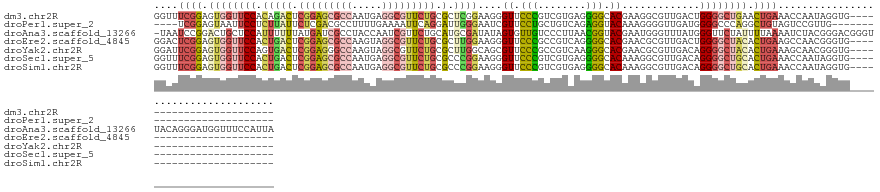
>dm3.chr2R 19943603 116 - 21146708 GGUUUCGGAGUGGUUCCACAGACUCGGAGCGCCAAUGAGGCGUUCUGCGCUCGGAAGGGUUCCCGUCGUGAGGGGCACGAAGGCGUUGACUGGGGCUGAACUGAAACCAAUAGGUG------------------------ ((((((((..((((((((..(((.(((((((((.....))))))))).((((....))))..((.(((((.....))))).)).)))...))))))))..))))))))........------------------------ ( -50.90, z-score = -2.56, R) >droPer1.super_2 7831251 109 - 9036312 ----UCGGAGUAAUUCCUCUUAUUCUCGACGCCUUUUGAAAAUUCAGGAUUGGGAAUCGUUCCUGCUGUCAGAGGUACAAAGGGGUUGAUGGGGCCCAGGCUGUAGUCCGUUG--------------------------- ----.((((.((...(((....((((((((.(((((((....(((..(((.((((.....))))...))).)))...)))))))))))).)))....)))...)).))))...--------------------------- ( -30.70, z-score = 0.33, R) >droAna3.scaffold_13266 5426848 139 + 19884421 -UAAUCCGGACUGCUCCAUUUUUUAUGAUCGCCUACCAAUCGUUCUGCAUGCGAUAUAGUGUUGUCCCUUAACGGUACGAAUGGGUUUAUGGGUUCUAUUUUAAAAUCUACGGGACGGGUUACAGGGAUGGUUUCCAUUA -(((((((..(((....(((((........((((((((.((((.(((...(.((((......)))).)....))).)))).))).....)))))........)))))...)))..)))))))..((((....)))).... ( -32.59, z-score = 0.84, R) >droEre2.scaffold_4845 21318845 116 - 22589142 GGACUCGGAGUGGUUCCACUGACUCGGAGCGCCAAGUAGGCGUUCUGCGCUUGGAAGGGUUCCCGCCGUCAGGGGCACGAACGCGUUGACUGGGGCUACACUGAAGCCAACGGGUG------------------------ ((..((((.(((((((((..(((.(((((((((.....)))))))(((.(((((..((....))....))))).)))....)).)))...))))))))).))))..))........------------------------ ( -48.90, z-score = -0.81, R) >droYak2.chr2R 19922448 116 - 21139217 GGAUUCGGAGUGGUUCCAGUGACUCGGAGGGCCAAGUAGGCGUUCUGCGCUUGGCAGCGUUCCCGCCGUCAAGGGCACGAACGCGUUGACAGGGGCUACACUGAAAGCAACGGGUG------------------------ ...(((((.((((((((....(((.((....)).)))..(((((((((.((((((.(((....))).)))))).))).)))))).......)))))))).)))))...........------------------------ ( -48.60, z-score = -1.56, R) >droSec1.super_5 197013 116 - 5866729 GGUUUCGGAGUGGUUCCACUGACUCGGAGCGCCAAUGAGGCGUUCUGCGCCCGGAAGGGUUCCCGUCGUGAGGGGCACAAAGGCGUUGACAGGGGCUGCACUGAAACCAAUAGGUG------------------------ ((((((((.(..(((((...(((.(((((((((.....))))))))).((((....))))....)))((.((.(.(.....).).)).)).)))))..).))))))))........------------------------ ( -54.50, z-score = -3.14, R) >droSim1.chr2R 18490311 116 - 19596830 GGUUUCGGAGUGGUUCCACUGACUCGGAGCGCCAAUGAGGCGUUCUGCGCCCGGAAGGGUUCCCGUCGUGAGGGGCACAAAGGCGUUGACAGGGGCUGCACUGAAACCAAUAGGUG------------------------ ((((((((.(..(((((...(((.(((((((((.....))))))))).((((....))))....)))((.((.(.(.....).).)).)).)))))..).))))))))........------------------------ ( -54.50, z-score = -3.14, R) >consensus GGAUUCGGAGUGGUUCCACUGACUCGGAGCGCCAAUGAGGCGUUCUGCGCUCGGAAGGGUUCCCGCCGUCAGGGGCACGAAGGCGUUGACAGGGGCUACACUGAAACCAAUAGGUG________________________ ....((((.((((((((.(((((.(((((((((.....))))))))).).))))....((.(((........))).)).............)))))))).)))).................................... (-23.95 = -25.27 + 1.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:14 2011