
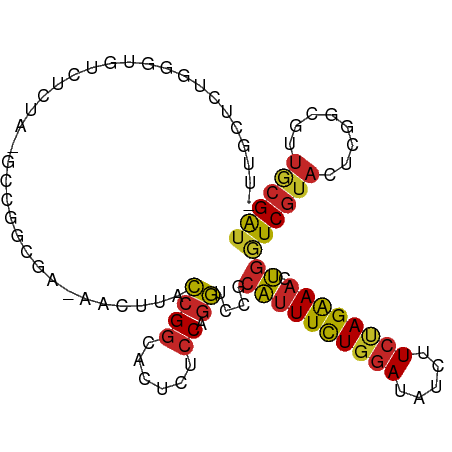
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,932,480 – 19,932,578 |
| Length | 98 |
| Max. P | 0.804013 |

| Location | 19,932,480 – 19,932,578 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 71.27 |
| Shannon entropy | 0.62686 |
| G+C content | 0.49416 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -10.98 |
| Energy contribution | -11.11 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19932480 98 + 21146708 AUGGGUCUGGGUGUCUCUA-ACCGGCGG-AACUUACCGGCACUCUCCAGGUCCGCAUUUCUGGAUAUCUUCUAGAAACUGGUCGUACUCGGCGUUGCGAU ..((..((((((.......-)))(.(((-......))).)......)))..))(((((((((((.....))))))))...((((....))))..)))... ( -33.30, z-score = -1.57, R) >droSim1.chr2R 18468772 98 + 19596830 AGUGCUCUGGGUGUCUCUA-GCCAGCGG-UACUUACCGGCACUCUCCAGGUCCGCAUUUCUGGAUAUCUUCUAGAAACUGGUCGUACUCGGCGUUGCGAU (((((.((((.((....))-.))))(((-(....))))))))).........((((((((((((.....))))))))...((((....))))..)))).. ( -31.60, z-score = -1.04, R) >droSec1.super_5 185894 98 + 5866729 AUUGCUCUGGGUGUCUCUA-GCCGGUGG-UACUUACCGGCACUCUCCAGGUCCGCAUUUCUGGAUAUCUUCUAGAAACUGGUCGUACUCGGCGUUGCGAU ...((.(((((.((.....-(((((((.-....))))))))).).)))))).((((((((((((.....))))))))...((((....))))..)))).. ( -33.40, z-score = -1.97, R) >droYak2.chr2R 19911239 99 + 21139217 UUUUCUUUAGGGGUCUCCAAGCUGGCGA-AACUUACCGGCACUCUCCGGGUCCGCAUUUCUGGAUAUCUUCUAGAAACUGGUCGUACUCGGCGUUGCGAU .......((((..((.((.....)).))-..))))...(((..(.((((((.(((.((((((((.....))))))))...).)).)))))).).)))... ( -29.70, z-score = -0.99, R) >droEre2.scaffold_4845 21307922 99 + 22589142 UUUGCCCUGAGGGUCUCUAAGCCCGCGG-AACUUACCGGCACUCUCCAGGUCCGCAUUUCUGGAUAUCUUCUAGAAACUGGUCGUACUCGGCGUUGCGAU .....((((.((((......))))((((-......)).))......))))..((((((((((((.....))))))))...((((....))))..)))).. ( -30.60, z-score = -0.64, R) >droAna3.scaffold_13266 5415727 98 - 19884421 --UGGGAUAUAUAACCAUAUAGUUAUGAUGACUUACCGGCACUCGCCGGGUCCGCACUUCUGGAUAUCUUCCAGAAACUGGUCGUAUUCGGCGUUGCGGU --((((.((((((((......))))).))).))))(((((....)))))..(((((.(((((((.....)))))))....((((....))))..))))). ( -33.90, z-score = -2.22, R) >dp4.chr3 7620285 95 + 19779522 -AUGGUAUAGGG---UAUAUGGAAGUAA-AACUUACCGGCAUUCGCCAGGACCGCAUUUCUGGAUAUCUUCUAGAAACUGGUCGUAUUCGCUGUUGCGAU -..((((.((..---(((......))).-..))))))(((....)))..(((((..((((((((.....)))))))).)))))....((((....)))). ( -28.80, z-score = -1.92, R) >droPer1.super_2 7815932 75 + 9036312 ------------------------GUAA-AACUUACCGGCAUUCGCCAGGACCGCAUUUCUGGAUAUCUUCUAGAAACUGGUCGUAUUCGCUGUUGCGAU ------------------------....-....(((.(((....)))..(((((..((((((((.....)))))))).)))))))).((((....)))). ( -24.40, z-score = -3.20, R) >droMoj3.scaffold_6496 12169410 74 - 26866924 -------------------------CAG-AACUUACCGGCAUUCUCCAGGACCGCAUUUCUGGAUAUCUUCUAGAAACUGAUCGUAUUCGCUAUUCCGAU -------------------------...-........((......)).(((..((.((((((((.....))))))))..((......))))...)))... ( -13.20, z-score = -0.14, R) >apiMel3.Group2 2839598 99 + 14465785 AUUGAUGAGUGUAUUCAAA-AUCGAAAAGGCCAUAGCGACAUUCUCCGCUACCACCUUUUUGCAAAUCUUCAAGGAAGGCAUCAUAUGCUGCAUCACGAG ..((((((((((((.....-..((((((((...(((((........)))))...))))))))....(((((...)))))....))))))).))))).... ( -25.10, z-score = -2.00, R) >consensus _UUGCUCUGGGUGUCUCUA_GCCGGCGA_AACUUACCGGCACUCUCCAGGUCCGCAUUUCUGGAUAUCUUCUAGAAACUGGUCGUACUCGGCGUUGCGAU ...................................((((......)).))....((((((((((.....)))))))).))((((((........)))))) (-10.98 = -11.11 + 0.13)


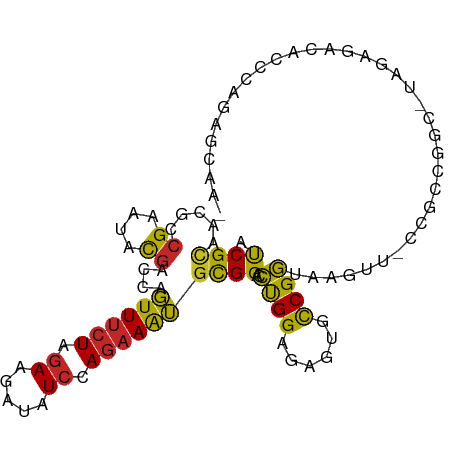
| Location | 19,932,480 – 19,932,578 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 71.27 |
| Shannon entropy | 0.62686 |
| G+C content | 0.49416 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -10.89 |
| Energy contribution | -10.29 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.601221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19932480 98 - 21146708 AUCGCAACGCCGAGUACGACCAGUUUCUAGAAGAUAUCCAGAAAUGCGGACCUGGAGAGUGCCGGUAAGUU-CCGCCGGU-UAGAGACACCCAGACCCAU ...((.((.....)).......((((((.((.....)).))))))))((..((((.(.((((((((.....-..))))))-.....)).)))))..)).. ( -22.60, z-score = -0.33, R) >droSim1.chr2R 18468772 98 - 19596830 AUCGCAACGCCGAGUACGACCAGUUUCUAGAAGAUAUCCAGAAAUGCGGACCUGGAGAGUGCCGGUAAGUA-CCGCUGGC-UAGAGACACCCAGAGCACU ...((.((.....)).......((((((((.......((((...(((..(((.((......)))))..)))-...)))))-))))))).......))... ( -24.01, z-score = -0.12, R) >droSec1.super_5 185894 98 - 5866729 AUCGCAACGCCGAGUACGACCAGUUUCUAGAAGAUAUCCAGAAAUGCGGACCUGGAGAGUGCCGGUAAGUA-CCACCGGC-UAGAGACACCCAGAGCAAU ...((.((.....)).......(((((((.......(((((..........)))))....((((((.....-..))))))-))))))).......))... ( -25.30, z-score = -1.53, R) >droYak2.chr2R 19911239 99 - 21139217 AUCGCAACGCCGAGUACGACCAGUUUCUAGAAGAUAUCCAGAAAUGCGGACCCGGAGAGUGCCGGUAAGUU-UCGCCAGCUUGGAGACCCCUAAAGAAAA .......(.((((((.(((((.((((((.((.....)).))))))..))..((((......))))......-)))...)))))).).............. ( -22.30, z-score = -0.06, R) >droEre2.scaffold_4845 21307922 99 - 22589142 AUCGCAACGCCGAGUACGACCAGUUUCUAGAAGAUAUCCAGAAAUGCGGACCUGGAGAGUGCCGGUAAGUU-CCGCGGGCUUAGAGACCCUCAGGGCAAA .(((((((((((.((((..(((((((((.((.....)).))))).......))))...))))))))..)))-..))))(((..(((...)))..)))... ( -27.11, z-score = 0.26, R) >droAna3.scaffold_13266 5415727 98 + 19884421 ACCGCAACGCCGAAUACGACCAGUUUCUGGAAGAUAUCCAGAAGUGCGGACCCGGCGAGUGCCGGUAAGUCAUCAUAACUAUAUGGUUAUAUAUCCCA-- .(((((....((....)).....((((((((.....)))))))))))))..(((((....))))).........((((((....))))))........-- ( -28.60, z-score = -1.76, R) >dp4.chr3 7620285 95 - 19779522 AUCGCAACAGCGAAUACGACCAGUUUCUAGAAGAUAUCCAGAAAUGCGGUCCUGGCGAAUGCCGGUAAGUU-UUACUUCCAUAUA---CCCUAUACCAU- .((((....))))....((((.((((((.((.....)).))))))..))))..(((....)))((.(((..-...))))).....---...........- ( -23.10, z-score = -2.14, R) >droPer1.super_2 7815932 75 - 9036312 AUCGCAACAGCGAAUACGACCAGUUUCUAGAAGAUAUCCAGAAAUGCGGUCCUGGCGAAUGCCGGUAAGUU-UUAC------------------------ .((((....))))....((((.((((((.((.....)).))))))..))))(((((....)))))......-....------------------------ ( -22.60, z-score = -2.71, R) >droMoj3.scaffold_6496 12169410 74 + 26866924 AUCGGAAUAGCGAAUACGAUCAGUUUCUAGAAGAUAUCCAGAAAUGCGGUCCUGGAGAAUGCCGGUAAGUU-CUG------------------------- ..((((((.........((((.((((((.((.....)).))))))..))))((((......))))...)))-)))------------------------- ( -16.50, z-score = -0.30, R) >apiMel3.Group2 2839598 99 - 14465785 CUCGUGAUGCAGCAUAUGAUGCCUUCCUUGAAGAUUUGCAAAAAGGUGGUAGCGGAGAAUGUCGCUAUGGCCUUUUCGAU-UUUGAAUACACUCAUCAAU ...(((.((((((((...))))((((...))))...))))(((((((.(((((((......))))))).)))))))....-........)))........ ( -27.30, z-score = -1.35, R) >consensus AUCGCAACGCCGAAUACGACCAGUUUCUAGAAGAUAUCCAGAAAUGCGGACCUGGAGAGUGCCGGUAAGUU_CCGCCGGC_UAGAGACACCCAGAGCAA_ .((((.....((....))....((((((.((.....)).))))))))))..((((......))))................................... (-10.89 = -10.29 + -0.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:14 2011