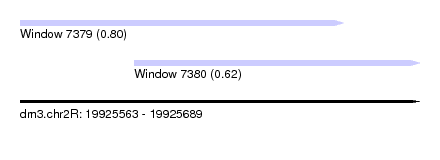
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,925,563 – 19,925,689 |
| Length | 126 |
| Max. P | 0.797677 |

| Location | 19,925,563 – 19,925,665 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 62.03 |
| Shannon entropy | 0.83273 |
| G+C content | 0.48054 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -10.33 |
| Energy contribution | -9.51 |
| Covariance contribution | -0.82 |
| Combinations/Pair | 2.25 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19925563 102 + 21146708 UUCACACACGAACAGAG-AA---AAAUGGAUCGCGGGAAAGGAAGCGGCUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGCAAG--- .................-..---...((((((((..........)))).)))).((((.((((((.((...((((((...))))))...)))))))).))))....--- ( -31.50, z-score = -0.64, R) >droVir3.scaffold_13324 2121154 94 + 2960039 ---------------AAAAAUAAGUAUGGAACGUAAUAAAACGAAUGUCCCUAAUAGGGGUCAAAAAAAAGUAUUACUGGGUGCAGAACUUUUUGCUCUUGGUACUAAA ---------------...........(((..(((......)))..((.((((....)))).))...........(((..((.(((((....))))).))..)))))).. ( -20.30, z-score = -1.93, R) >droGri2.scaffold_15245 6424109 107 - 18325388 --UUUAGUAAAUAACAGAGACCAAAAUGGAUCGUAACAAAUCAAGCGCCGUCAAUAGGGGUCAGAAAAAGGCACUAUUAGGUGCCGAAUUAUUUGCACUUGGUACCAAA --....((((((((....((((......((((((..........)))..)))......)))).......((((((....))))))...))))))))..(((....))). ( -25.50, z-score = -1.99, R) >droPer1.super_2 7808608 102 + 9036312 ---UCCUCAUAAUAAAG-UAAAAAAAUGGAUCGCGGAAAAGGUGG---AUCGGGUAGAUCACAGAAAAAGGUGCCACUUGGUGGGGAACUGUUUGCGCUUGGAACAAAG ---(((........(((-(....((((((.((.(.(...((((((---(((.....)))(((........)))))))))..).).)).))))))..)))))))...... ( -18.20, z-score = 0.86, R) >dp4.chr3 7612973 98 + 19779522 -------CAUAAUAAAG-UAAAAAAAUGGAUCGCGGAAAAGGUGG---AUCGGGUAGAUCACAGAAAAAGGUGCCACUUGGUGGGGAACUGUUUGCGCUUGGAACAAAG -------.......(((-(....((((((.((.(.(...((((((---(((.....)))(((........)))))))))..).).)).))))))..))))......... ( -17.90, z-score = -0.36, R) >droAna3.scaffold_13266 5408680 108 - 19884421 AUAAUUCCAGAAAAUAA-AAGUGGAAUGGAUCGCGGUAAAGGCGGCGGAUCCAAUAGAUCCCAGAAAAAAGUGCCUCUGGGCGGCGAACUCUUUGCCCUCGGCACCAAA ...((((((........-...))))))(((((((.((....)).)).)))))..................(((((...((((((.(....).))))))..))))).... ( -33.40, z-score = -1.17, R) >droEre2.scaffold_4845 21301015 105 + 22589142 UUAAUUUGCGAACAAAG-ACACUACAUGGAUCGCGGGAAAGGAAGCGGGUCGAACCGGUCGCAAAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGCAAG--- ....(((((((.....(-((.((....)).((((..........))))))).......)))))))..........((((((((.(((....))).))))))))...--- ( -30.50, z-score = -0.14, R) >droYak2.chr2R 19904315 106 + 21139217 UUAAUUUGCGAACAGAUUACACUAAAUGGAUCGCGGGAAAGGAAGCGGAUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUCGCGCUCGGCAAG--- .(((((((....))))))).......((((((((..........)).)))))).((((.(((.(((((...((((((...))))))...)))))))).))))....--- ( -36.80, z-score = -2.19, R) >droSec1.super_5 178751 105 + 5866729 UUCAUACUCGAAAAAAG-ACACUAAAUGGAUCGCGGGAAAGGAAGCGGCUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGCAAG--- .................-........((((((((..........)))).)))).((((.((((((.((...((((((...))))))...)))))))).))))....--- ( -31.50, z-score = -0.54, R) >droSim1.chr2R 18462223 103 + 19596830 UUCAUACUCUAACAAAG-ACACUAAAUGGAUCGCGGGAAAG-AAGCGGCUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUU-GCGCUCGGCAAG--- .................-........((((((((.......-..)))).)))).((((.((((..((....((((((...))))))..))..)-))).))))....--- ( -29.90, z-score = -0.63, R) >droMoj3.scaffold_6496 14865467 84 + 26866924 -------------------------AUGGAUCGAAAUAAAACAAAUGCCCCGAAUAGGGGCCAAAAGAAAGUAUUGCUGGGUGCCGAACUCUUUGCUCUCGGUACCAAA -------------------------.((((((((............(((((.....)))))..............((.((((.....))))...))..))))).))).. ( -23.70, z-score = -1.64, R) >droWil1.scaffold_180700 5041631 84 + 6630534 -------------------------AUGGAGCGCUCCAAAAGUAGCGGCUCAAAUAGGGGCCAAAAGAAAGUGCCGCUUGGUGGCGAACUCUUUGCACUGGGCACAAAA -------------------------.....(.((((....(((.(((((((......))))).(((((...((((((...))))))...)))))))))))))).).... ( -27.40, z-score = -0.69, R) >consensus ___AU___CGAACAAAG_AAACAAAAUGGAUCGCGGGAAAGGAAGCGGCUCCAAUAGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGCACGAA_ ..................................((((.........((((.....))))...........))))((((((((............))))))))...... (-10.33 = -9.51 + -0.82)


| Location | 19,925,599 – 19,925,689 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 70.52 |
| Shannon entropy | 0.63334 |
| G+C content | 0.53916 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -12.80 |
| Energy contribution | -11.94 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 2.13 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.624329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19925599 90 + 21146708 GGAAGCGGCUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGCA---AGAGCGUACGCGAUGACUCCAAGUCG ....(((((((...((((.((((((.((...((((((...))))))...)))))))).))))..---.))))...)))...(((.....))). ( -34.40, z-score = -0.87, R) >droVir3.scaffold_13324 2121179 93 + 2960039 ACGAAUGUCCCUAAUAGGGGUCAAAAAAAAGUAUUACUGGGUGCAGAACUUUUUGCUCUUGGUACUAAAAGUUGUCGUGACGAUUCUAAGACG ..((((..(((.....)))((((..((....((.(((..((.(((((....))))).))..))).))....))....)))).))))....... ( -22.70, z-score = -1.47, R) >droGri2.scaffold_15245 6424147 90 - 18325388 UCAAGCGCCGUCAAUAGGGGUCAGAAAAAGGCACUAUUAGGUGCCGAAUUAUUUGCACUUGGUACCAAAACUGGUCGCGAUGAUCCCAAG--- ................(((((((......((((((....)))))).......((((..(..((......))..)..)))))))))))...--- ( -26.90, z-score = -1.96, R) >droPer1.super_2 7808644 90 + 9036312 ---GGUGGAUCGGGUAGAUCACAGAAAAAGGUGCCACUUGGUGGGGAACUGUUUGCGCUUGGAACAAAGAGCUGUCGAGAUGAUCCAAAAUCC ---..(((((((......((((((......((.(((...((((.(((....))).))))))).))......)))).))..)))))))...... ( -22.10, z-score = 0.22, R) >dp4.chr3 7613005 90 + 19779522 ---GGUGGAUCGGGUAGAUCACAGAAAAAGGUGCCACUUGGUGGGGAACUGUUUGCGCUUGGAACAAAGAGCUGUCGAGAUGAUCCAAAAUCC ---..(((((((......((((((......((.(((...((((.(((....))).))))))).))......)))).))..)))))))...... ( -22.10, z-score = 0.22, R) >droAna3.scaffold_13266 5408719 93 - 19884421 GGCGGCGGAUCCAAUAGAUCCCAGAAAAAAGUGCCUCUGGGCGGCGAACUCUUUGCCCUCGGCACCAAAAGCGUCCGCGAUGACUCCAAGUCC .((((((((((.....))))).........(((((...((((((.(....).))))))..))))).......).))))...(((.....))). ( -32.70, z-score = -1.53, R) >droEre2.scaffold_4845 21301054 90 + 22589142 GGAAGCGGGUCGAACCGGUCGCAAAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGCA---AGAGCGUUCGCGAUGACUCCAAGUCA (((..(((......)))(((((....(((.((((.((((((((.(((....))).)))))))).---.)))).))))))))...)))...... ( -35.60, z-score = -0.99, R) >droYak2.chr2R 19904355 90 + 21139217 GGAAGCGGAUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUCGCGCUCGGCA---AGAGCGUACGCGAUGACUCCAAAUCA (((..(((......)))(((((.(((((...((((((...))))))...)))))((((((....---.))))))..)))))...)))...... ( -36.90, z-score = -2.28, R) >droSec1.super_5 178790 90 + 5866729 GGAAGCGGCUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGCA---AGAGCGUACGCGAUGAUUCCAAGUCG ((((.(((......)))(((((.(((((...((((((...))))))...)))))((((((....---.))))))..)))))..))))...... ( -35.40, z-score = -1.35, R) >droSim1.chr2R 18462262 88 + 19596830 -GAAGCGGCUCCAACCGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUU-GCGCUCGGCA---AGAGCGUACGCGAUGACUCCAAGUCG -...(((((((...((((.((((..((....((((((...))))))..))..)-))).))))..---.))))...)))...(((.....))). ( -32.70, z-score = -0.66, R) >droMoj3.scaffold_6496 14865482 93 + 26866924 ACAAAUGCCCCGAAUAGGGGCCAAAAGAAAGUAUUGCUGGGUGCCGAACUCUUUGCUCUCGGUACCAAAAGUGGCCGCGAUGAUUCUAAAGGA ......(((((.....)))))....((((..((((((..((((((((...........))))))))....(....))))))).))))...... ( -28.60, z-score = -1.16, R) >droWil1.scaffold_180700 5041646 93 + 6630534 AGUAGCGGCUCAAAUAGGGGCCAAAAGAAAGUGCCGCUUGGUGGCGAACUCUUUGCACUGGGCACAAAAAGUGUUCGCGAUGAUUCAAAAUCC (((.(((((((......))))).(((((...((((((...))))))...))))))))))((((((.....))))))..(((........))). ( -29.20, z-score = -1.38, R) >consensus GGAAGCGGCUCCAAUAGAUCGCAGAAGAAGGUUCCGCUGGGCGGAGAACUCUUUGCGCUCGGCAC_AAGAGCGUUCGCGAUGAUUCCAAGUCC ....(((((((.....))))...............((((((((............))))))))............)))............... (-12.80 = -11.94 + -0.86)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:12 2011