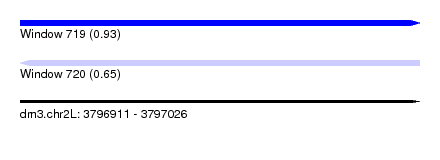
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,796,911 – 3,797,026 |
| Length | 115 |
| Max. P | 0.932878 |

| Location | 3,796,911 – 3,797,026 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.23 |
| Shannon entropy | 0.40019 |
| G+C content | 0.41438 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -18.37 |
| Energy contribution | -19.37 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
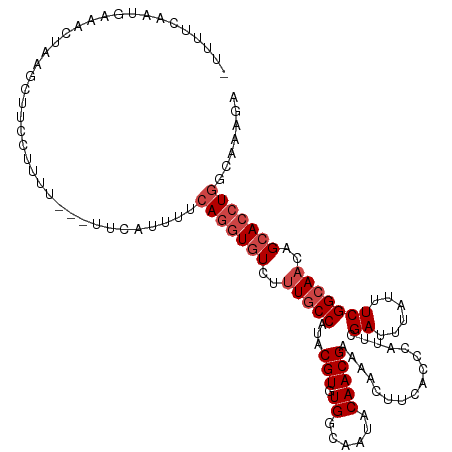
>dm3.chr2L 3796911 115 + 23011544 -UUUUCAAUGUAGCUAAGCUUUCUUUU---UGCAUUUUAAGGUAUCUUUGCCAUACGUGUGGCAAUACAACGAAAACUUCACGCAUUCGAUUUAUUUCGGCAACAGCACCUGGCAAAGA -.....(((((((..(((....))).)---))))))........(((((((((...((((.((........((.....)).......(((......)))))....)))).))))))))) ( -23.30, z-score = -0.56, R) >droSim1.chr2L 3752947 112 + 22036055 UUUUCUAGCUAACCU----UUCUUUUU---UUCCUUUUCAGGUGUCUUUGCCAUACGUGUGGCAAUACAACGAAAACUUCACCCAUUCGAUUUAUUUCGGCAACAGCACCUGGCAAAGA ...............----........---...((((((((((((..(((((....((((....))))...((((...((........))....)))))))))..)))))))).)))). ( -27.20, z-score = -3.20, R) >droSec1.super_5 1898995 118 + 5866729 -UUUUCAAUGUAGCUAAGCUUUCUUUUCCUUUCAUUUUCAGGUGUCUUUGCCAUACGUGUGGCAAUACAACGAAAACUUCACCCAUUCGAUUUAUUUCGGCAACAGCACCUGGCAAAGA -.......((((((...)))..................(((((((..(((((....((((....))))...((((...((........))....)))))))))..)))))))))).... ( -25.10, z-score = -1.53, R) >droYak2.chr2L 3798616 116 + 22324452 UUCUCUGCUAAAACUAAGCUUAAUGUU---UCCAUUUUCAGGUGUCUUUGCCAUACGUGUGGCAGUACAACGAAAACUUCACACAUUCGAUUUAUUUCGGCAACAGCACCUGGCAAAGA .(((.(((.......((((.....)))---).......(((((((..(((((....((((((.(((.........)))))))))....((......)))))))..)))))))))).))) ( -33.70, z-score = -3.83, R) >droEre2.scaffold_4929 3842393 113 + 26641161 ----UUCAUGCAAUGUAACUAGCUUUU--UUCCAUUUUCAGAUGUCUUUGCCAUACGUGUGGCAGUACAACGAAAACUUCACUCACUCGAUUUACUUCGGCAACAGCACGUGGCAAAGA ----.......................--...((((....))))((((((((..((((((.((........((.....)).......(((......)))))....)))))))))))))) ( -23.00, z-score = -0.33, R) >droAna3.scaffold_12916 1802584 114 + 16180835 ----UCUUCAAGAUUAUACAUCGUUCCA-AUUCCUCCACAGGUGUCCCUUCCAUACGUCUGGCAGUACAACGAAAACUUCACCCACUCGAUUUACUUCGGCAACAGCACCUGGCAGAGG ----........................-...((((..(((((((.....((...(((.((......)))))................((......)))).....)))))))...)))) ( -21.00, z-score = -0.67, R) >consensus _UUUUCAAUGAAACUAAGCUUCCUUUU___UUCAUUUUCAGGUGUCUUUGCCAUACGUGUGGCAAUACAACGAAAACUUCACCCAUUCGAUUUAUUUCGGCAACAGCACCUGGCAAAGA ......................................(((((((..(((((...(((.((......)))))................((......)))))))..)))))))....... (-18.37 = -19.37 + 1.00)

| Location | 3,796,911 – 3,797,026 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.23 |
| Shannon entropy | 0.40019 |
| G+C content | 0.41438 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -20.57 |
| Energy contribution | -20.27 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
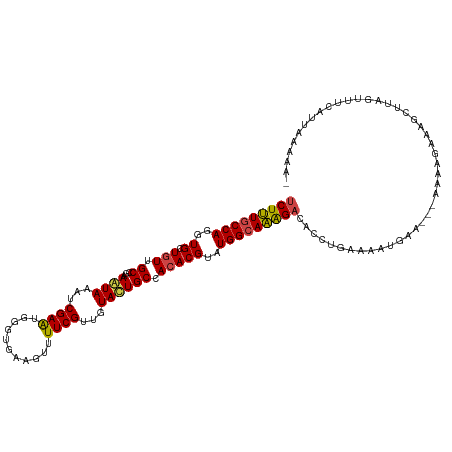
>dm3.chr2L 3796911 115 - 23011544 UCUUUGCCAGGUGCUGUUGCCGAAAUAAAUCGAAUGCGUGAAGUUUUCGUUGUAUUGCCACACGUAUGGCAAAGAUACCUUAAAAUGCA---AAAAGAAAGCUUAGCUACAUUGAAAA- .........(((......)))........((((..((...((((((((.(((((((((((......)))).(((....)))..))))))---)...)))))))).))....))))...- ( -26.00, z-score = -0.80, R) >droSim1.chr2L 3752947 112 - 22036055 UCUUUGCCAGGUGCUGUUGCCGAAAUAAAUCGAAUGGGUGAAGUUUUCGUUGUAUUGCCACACGUAUGGCAAAGACACCUGAAAAGGAA---AAAAAGAA----AGGUUAGCUAGAAAA .(((..((.(((......)))((......)).....))..)))((((((((...((((((......)))))).))).(((....)))..---.....)))----))............. ( -26.20, z-score = -0.85, R) >droSec1.super_5 1898995 118 - 5866729 UCUUUGCCAGGUGCUGUUGCCGAAAUAAAUCGAAUGGGUGAAGUUUUCGUUGUAUUGCCACACGUAUGGCAAAGACACCUGAAAAUGAAAGGAAAAGAAAGCUUAGCUACAUUGAAAA- .........(((......)))........((((...(((.(((((((((((...((((((......)))))).))).(((.........)))....)))))))).)))...))))...- ( -27.90, z-score = -0.96, R) >droYak2.chr2L 3798616 116 - 22324452 UCUUUGCCAGGUGCUGUUGCCGAAAUAAAUCGAAUGUGUGAAGUUUUCGUUGUACUGCCACACGUAUGGCAAAGACACCUGAAAAUGGA---AACAUUAAGCUUAGUUUUAGCAGAGAA (((((((((((((((.(((((((......))..(((((((.(((.........)))..)))))))..))))))).))))))..((((..---..)))).............))))))). ( -40.30, z-score = -4.51, R) >droEre2.scaffold_4929 3842393 113 - 26641161 UCUUUGCCACGUGCUGUUGCCGAAGUAAAUCGAGUGAGUGAAGUUUUCGUUGUACUGCCACACGUAUGGCAAAGACAUCUGAAAAUGGAA--AAAAGCUAGUUACAUUGCAUGAA---- (((((((((..((.(((.((...((((...((((...........))))...)))))).)))))..)))))))))..((....((((...--............))))....)).---- ( -25.76, z-score = 0.05, R) >droAna3.scaffold_12916 1802584 114 - 16180835 CCUCUGCCAGGUGCUGUUGCCGAAGUAAAUCGAGUGGGUGAAGUUUUCGUUGUACUGCCAGACGUAUGGAAGGGACACCUGUGGAGGAAU-UGGAACGAUGUAUAAUCUUGAAGA---- ((((..(.(((((((.((.....((((...((((...........))))...)))).(((......))))).)).))))))..))))...-......(((.....))).......---- ( -28.60, z-score = 0.14, R) >consensus UCUUUGCCAGGUGCUGUUGCCGAAAUAAAUCGAAUGGGUGAAGUUUUCGUUGUACUGCCACACGUAUGGCAAAGACACCUGAAAAUGAA___AAAAGAAAGCUUAGUUUCAUUAAAAA_ (((((((((..((.(((.((...((((...((((...........))))...)))))).)))))..)))))))))............................................ (-20.57 = -20.27 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:41 2011