
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,895,728 – 19,895,820 |
| Length | 92 |
| Max. P | 0.657709 |

| Location | 19,895,728 – 19,895,820 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 48.23 |
| Shannon entropy | 0.94516 |
| G+C content | 0.38409 |
| Mean single sequence MFE | -27.19 |
| Consensus MFE | -2.10 |
| Energy contribution | -0.63 |
| Covariance contribution | -1.47 |
| Combinations/Pair | 2.44 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.08 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.657709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19895728 92 - 21146708 -----------------GCAAGUUCCUCUUUAGAAUGAGAAUUGGUUGUCAACGAGGGGUUCUUGGA---AUCUCU---GGAUUCUCGGGAUUUCAGGAAUGCCUCUGUCUAAAA--- -----------------...........((((((.(((.((....)).)))..((((.(((((((((---(((.((---((....)))))))))))))))).))))..)))))).--- ( -31.70, z-score = -2.56, R) >droWil1.scaffold_180700 1543536 81 + 6630534 --------------------------GGACCGAAUUUGGCAUUUAAUGUGACGAAAUUAUAUACGAA---AUUUUUUUAGUUUGUGCCUGUUUUUUCAAUCGAUGUUAAA-------- --------------------------........(((((((((.....(((.(((((....((((((---..........))))))...))))).)))...)))))))))-------- ( -11.00, z-score = 0.45, R) >droPer1.super_2 7773530 108 - 9036312 GUCAGUUUUCCAAUUGGUUGCAAGUCAAUUCAGAUU-UUAAAGAAAAACGAUGCAAAAAUAUAUGUACUAAUCUUUGACGAGUGUGCAAAUAUUUUUCAUUG-UCUUAAA-------- .((.((...((....))..))(((((......))))-)....))...((((((.((((((((.(((((...((......))..))))).)))))))))))))-)......-------- ( -20.80, z-score = -1.43, R) >dp4.chr3 7577999 109 - 19779522 GUCAGUUUUCCAAUUGGUUGCAAGUCAAUUCAGAUUAAAAAAAAAAAACGAUGCAAAAAUAUAUGUAUUCAUCUUUUACGAGUGUGCAUAUAUUUUUCAUUG-UCUUAAA-------- ....((...((....))..)).((((......))))...........((((((.((((((((((((((...((......))..)))))))))))))))))))-)......-------- ( -22.90, z-score = -3.20, R) >droAna3.scaffold_13266 5766147 92 - 19884421 ---------------------GCUCGUUCCUUGAUUCA-AAUUGGUUAUU-CCGAAAACUUCUUAAA---GUCUCUACGAAAAUCUAGAGGUUUUUCGAAUGUCAAAAAACAAAACAC ---------------------....(((..(((((...-..((((.....-))))....(((..(((---..(((((.(.....))))))..)))..))).)))))..)))....... ( -16.30, z-score = -1.48, R) >droEre2.scaffold_4845 21268839 92 - 22589142 -----------------GCUAGUUCCUGUUUAGAAUGAGAAUUGGUUUCCAGGGAGGGGUUCCUGGA---GUCUCU---GGACUCUCGAGAUUUCAGGAACGCCUCCGUCUAAAA--- -----------------((((((((..((.....))..))))))))......(((((.(((((((((---(((((.---((...)).)))))))))))))).)))))........--- ( -40.70, z-score = -3.71, R) >droYak2.chr2R 19874152 92 - 21139217 -----------------GCUAGUUCCUGUUCAGAAUGAGAAUUGGUUUCCGAAGAGGGGUUCCUGGA---AUCUCU---GGACUCUCGAGAUCUCAGGAACACCUCAGUCUAAAA--- -----------------((((((((..((.....))..)))))))).......((((.(((((((..---(((((.---((...)).)))))..))))))).)))).........--- ( -32.90, z-score = -1.76, R) >droSec1.super_9 3179704 91 - 3197100 -----------------GCAAGUUCCUGUUUAGAAUGAGAAUUGGUUGUCAACGAGGGGUUCUUGGA---AUCUCU---GGA-UCUCGAGAUUUCAGGAACGCCUCUGUCUAAAA--- -----------------...........((((((.(((.((....)).)))..((((.(((((((((---(((((.---...-....)))))))))))))).))))..)))))).--- ( -34.20, z-score = -3.55, R) >droSim1.chr2R 18441600 91 - 19596830 -----------------GCAAGUUCCUGUUUAGAAUGAGAAUUGGUUGUCAACGAGGGGUUCUUGGA---AUCUCU---GGA-UCUCGAGAUUUCAGGAACGCCUCUGUCUAAAA--- -----------------...........((((((.(((.((....)).)))..((((.(((((((((---(((((.---...-....)))))))))))))).))))..)))))).--- ( -34.20, z-score = -3.55, R) >consensus _________________GCAAGUUCCUGUUUAGAAUGAGAAUUGGUUGUCAACGAGGGGUUCUUGGA___AUCUCU___GGAUUCUCGAGAUUUCAGGAACGCCUCUGUCUAAAA___ ........................................................(((.((((((...................)))))).)))....................... ( -2.10 = -0.63 + -1.47)

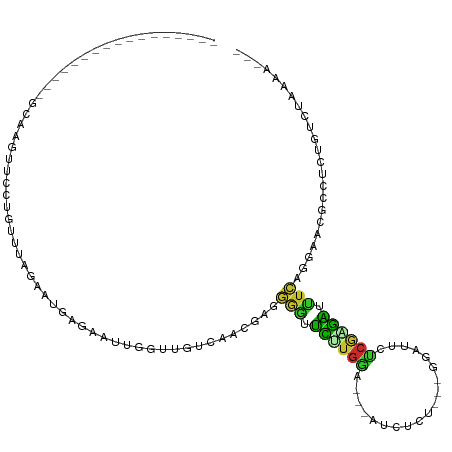
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:06 2011