| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,844,964 – 19,845,060 |
| Length | 96 |
| Max. P | 0.682607 |

| Location | 19,844,964 – 19,845,060 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 71.35 |
| Shannon entropy | 0.44431 |
| G+C content | 0.40600 |
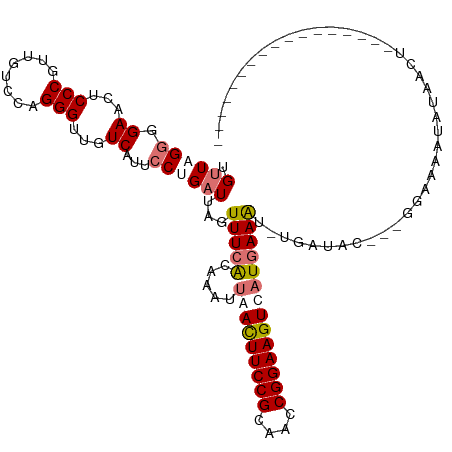
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -16.21 |
| Energy contribution | -17.65 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
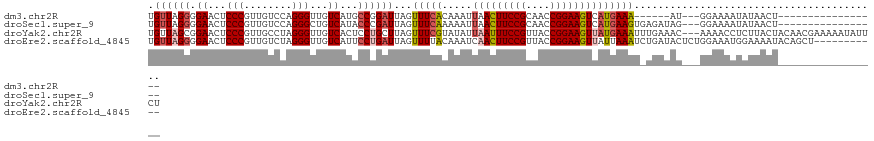
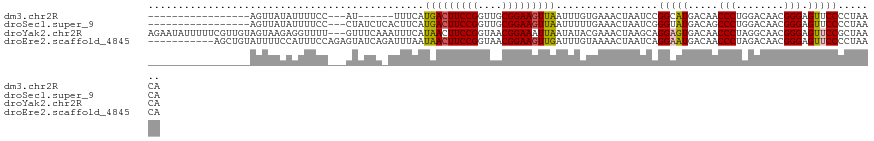
>dm3.chr2R 19844964 96 + 21146708 UGUUAGGGGAACUCCCGUUGUCCAGGGUUGUCAUGCCGGAUUAGUUUCACAAAUUAACUUCCGCAACCGGAAGUCAUGAAA------AU---GGAAAAUAUAACU----------------- .((((.(((....)))(((.((((......(((((..(((.....)))........(((((((....))))))))))))..------.)---))).))).)))).----------------- ( -24.40, z-score = -0.79, R) >droSec1.super_9 3128849 102 + 3197100 UGUUAGGGGAACUCCCGUUGUCCAGGGCUGUCAUACCCGAUUAGUUUCAAAAAUUAACUUCCGCAACCGGAAGUCAUGAAGUGAGAUAG---GGAAAAUAUAACU----------------- .((((.(((....)))(((.(((.(((........))).....((((((.......(((((((....))))))).......))))))..---))).))).)))).----------------- ( -27.84, z-score = -1.36, R) >droYak2.chr2R 19822311 119 + 21139217 UGUUAGCGGAACUCCCGUUGCCUAGGGUUGUCACUCCUGCUUAGUUUCGUAUAUUAAUUUCCGUUACCGGAAGUUAUGAAAUUUGAAAC---AAAACCUCUUACUACAACGAAAAAUAUUCU .(((((((((((.(((........)))..))...))).)))..((((((.....(((((((((....))))))))).......))))))---...............)))............ ( -24.90, z-score = -1.33, R) >droEre2.scaffold_4845 21217717 111 + 22589142 UGUUAGGGGAACUCCCGUUGUCUAGGGUUGUCAUUCCUGAUUAGUUUUACAAAUCAACUUCCGUUACCGGAAGUUAUUAAAUCUGAUACUCUGGAAAUGGAAAAUACAGCU----------- ......(((....)))((((((((((((.((((.....((((.(.....).))))((((((((....))))))))........))))))))))))..(....)...)))).----------- ( -29.90, z-score = -1.75, R) >consensus UGUUAGGGGAACUCCCGUUGUCCAGGGUUGUCAUUCCUGAUUAGUUUCACAAAUUAACUUCCGCAACCGGAAGUCAUGAAAU_UGAUAC___GGAAAAUAUAACU_________________ .((((((.((...(((........)))...))...))))))...(((((.....(((((((((....))))))))))))))......................................... (-16.21 = -17.65 + 1.44)
| Location | 19,844,964 – 19,845,060 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 71.35 |
| Shannon entropy | 0.44431 |
| G+C content | 0.40600 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -17.05 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19844964 96 - 21146708 -----------------AGUUAUAUUUUCC---AU------UUUCAUGACUUCCGGUUGCGGAAGUUAAUUUGUGAAACUAAUCCGGCAUGACAACCCUGGACAACGGGAGUUCCCCUAACA -----------------.((((.....(((---(.------((((((((((((((....)))))))).....)))))).......((........)).))))....(((....))).)))). ( -22.90, z-score = -0.72, R) >droSec1.super_9 3128849 102 - 3197100 -----------------AGUUAUAUUUUCC---CUAUCUCACUUCAUGACUUCCGGUUGCGGAAGUUAAUUUUUGAAACUAAUCGGGUAUGACAGCCCUGGACAACGGGAGUUCCCCUAACA -----------------.((((....((((---(....(((.....(((((((((....))))))))).....)))........((((......))))........)))))......)))). ( -26.70, z-score = -1.28, R) >droYak2.chr2R 19822311 119 - 21139217 AGAAUAUUUUUCGUUGUAGUAAGAGGUUUU---GUUUCAAAUUUCAUAACUUCCGGUAACGGAAAUUAAUAUACGAAACUAAGCAGGAGUGACAACCCUAGGCAACGGGAGUUCCGCUAACA .((((...(..((((((........((((.---(((((........(((.(((((....))))).)))......))))).))))(((.((....)))))..))))))..)))))........ ( -27.24, z-score = -1.15, R) >droEre2.scaffold_4845 21217717 111 - 22589142 -----------AGCUGUAUUUUCCAUUUCCAGAGUAUCAGAUUUAAUAACUUCCGGUAACGGAAGUUGAUUUGUAAAACUAAUCAGGAAUGACAACCCUAGACAACGGGAGUUCCCCUAACA -----------((((........((((((...(((..((((((....((((((((....))))))))))))))....))).....))))))....(((........)))))))......... ( -24.30, z-score = -1.01, R) >consensus _________________AGUUAUAUUUUCC___GUAUCA_AUUUCAUAACUUCCGGUAACGGAAGUUAAUUUGUGAAACUAAUCAGGAAUGACAACCCUAGACAACGGGAGUUCCCCUAACA ..............................................(((((((((....))))))))).................(((((.....(((........))).)))))....... (-17.05 = -17.18 + 0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:02 2011