| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,828,290 – 19,828,359 |
| Length | 69 |
| Max. P | 0.977005 |

| Location | 19,828,290 – 19,828,359 |
|---|---|
| Length | 69 |
| Sequences | 11 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 77.57 |
| Shannon entropy | 0.46947 |
| G+C content | 0.36220 |
| Mean single sequence MFE | -13.72 |
| Consensus MFE | -7.56 |
| Energy contribution | -7.84 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.977005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19828290 69 - 21146708 ACACACGUAAGCACCAAAGGUCAAACUAAAGUCA-------GCUCGGAUUU--GCUUUUCACUUUUAUUUGAUCUUCA ................(((((((((..(((((.(-------((........--)))....)))))..))))))))).. ( -13.70, z-score = -2.53, R) >droPer1.super_2 1958423 70 - 9036312 ACACACGUAUGCACAUAAGGUCAAACUAAAGUCA------GGUUGAAUUCU--GACUGUCACUUUUAUUUGAUCUUCA ................(((((((((..(((((..------((((.......--))))...)))))..))))))))).. ( -14.20, z-score = -1.43, R) >dp4.chr3 1768220 70 - 19779522 ACACACGUAUGCACAUAAGGUCAAACUAAAGUCA------GGUUGAAUUCU--GACUGUCACUUUUAUUUGAUCUUCA ................(((((((((..(((((..------((((.......--))))...)))))..))))))))).. ( -14.20, z-score = -1.43, R) >droAna3.scaffold_13266 5693022 70 - 19884421 ACACACGUAAGCACAUAAGGUCAAACUAAAGUCA------GGCUGGAAUUU--ACUUUUCACUUCUAUUUGAUCUUCA ................(((((((((....((..(------((.(((((...--...)))))))))).))))))))).. ( -8.70, z-score = -0.56, R) >droEre2.scaffold_4845 21200913 69 - 22589142 ACACACGUAAGCACCUAAGGUCAAACUAAAGUCA-------GCUCGGCUUU--GCUUUUCACUUUUAUUUGAUCUUCA ................(((((((((..(((((.(-------((........--)))....)))))..))))))))).. ( -13.60, z-score = -2.36, R) >droYak2.chr2R 19805743 69 - 21139217 ACACACGUAAGCACCUAAGGUCAAACUAAAGUCA-------GCUCGGCUUU--GCUUUUCACUUUUAUUUGAUCUUCA ................(((((((((..(((((.(-------((........--)))....)))))..))))))))).. ( -13.60, z-score = -2.36, R) >droSec1.super_9 3112308 69 - 3197100 ACACACGUAAGCACCUAAGGUCAAACUAAAGUCA-------GCUCGGCUUU--GCUUUUCACUUUUAUUUGAUCUUCA ................(((((((((..(((((.(-------((........--)))....)))))..))))))))).. ( -13.60, z-score = -2.36, R) >droSim1.chr2R 18380617 69 - 19596830 ACACACGUAAGCACCUAAGGUCAAACUAAAGUCA-------GCUCGGCUUU--GCUUUUCACUUUUAUUUGAUCUUCA ................(((((((((..(((((.(-------((........--)))....)))))..))))))))).. ( -13.60, z-score = -2.36, R) >droWil1.scaffold_180701 2749539 69 + 3904529 --------AUACA-AUAAGGUCAAAAUAAAGUCAGCUUGACGCUUCACUUUCAGGUUUUAACUUUUAUUUGAUCUUGA --------.....-.((((((((((..(((((.(((((((..........)))))))...)))))..)))))))))). ( -16.90, z-score = -3.62, R) >droMoj3.scaffold_6496 11934621 68 + 26866924 ACAAAAAUGAAAAAAGAGGGUCAAACUAAAGUCA-------GACUCACCUU---CGUCUAACUUUUAUUUGAACUUCA ...............((((.(((((..(((((.(-------(((.......---.)))).)))))..))))).)))). ( -17.40, z-score = -3.53, R) >droGri2.scaffold_15245 11775218 70 - 18325388 AUACAUUCACCUGAAUAAGGUCAAACAAAUGUCG-------ACCUCACUUUC-GACUUUAACUUUGAUUUGAACUUCA ............(((..((((((((.....((((-------(........))-)))......))))))))....))). ( -11.40, z-score = -1.91, R) >consensus ACACACGUAAGCACAUAAGGUCAAACUAAAGUCA_______GCUCGACUUU__GCUUUUCACUUUUAUUUGAUCUUCA ................(((((((((..(((((............................)))))..))))))))).. ( -7.56 = -7.84 + 0.28)

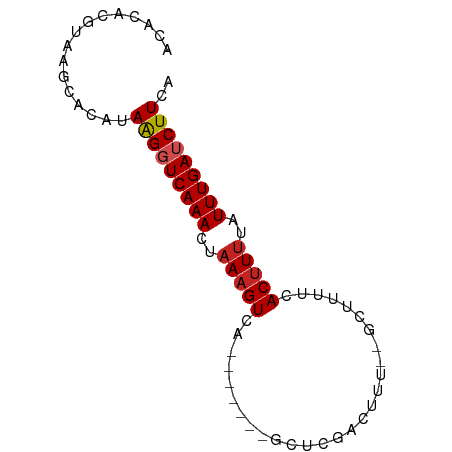
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:47:00 2011