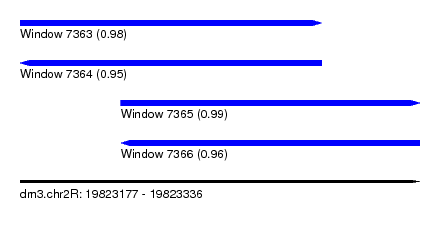
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,823,177 – 19,823,336 |
| Length | 159 |
| Max. P | 0.989685 |

| Location | 19,823,177 – 19,823,297 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.71 |
| Shannon entropy | 0.05077 |
| G+C content | 0.37292 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -23.84 |
| Energy contribution | -23.85 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19823177 120 + 21146708 CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGCUGCGCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUA (((.........(.(((..(....)))).)(((.(.....)..))))))..((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. ( -26.90, z-score = -2.63, R) >droSim1.chr2R 18375569 120 + 19596830 CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGCUGCGCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUA (((.........(.(((..(....)))).)(((.(.....)..))))))..((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. ( -26.90, z-score = -2.63, R) >droSec1.super_9 3107153 120 + 3197100 CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGCUGCGCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCCA (((.........(.(((..(....)))).)(((.(.....)..))))))..((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. ( -26.50, z-score = -2.41, R) >droYak2.chr2R 19800778 120 + 21139217 CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGCUGCUCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUA ...........(((....((((.......(....)......)))).)))..((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. ( -25.52, z-score = -2.57, R) >droEre2.scaffold_4845 21195937 120 + 22589142 CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGCUGCUCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUA ...........(((....((((.......(....)......)))).)))..((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. ( -25.52, z-score = -2.57, R) >droAna3.scaffold_13266 5688203 120 + 19884421 CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGCUGCUCGUUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUA .........(((((....((((.......(....)......)))).)))))((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. ( -27.42, z-score = -3.02, R) >dp4.chr3 1764625 120 + 19779522 CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGCUGCUUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUA .............(((((((((.......(....)......)))).)))))((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. ( -28.62, z-score = -3.68, R) >droPer1.super_2 1952213 120 + 9036312 CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGCUGCUUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUA .............(((((((((.......(....)......)))).)))))((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. ( -28.62, z-score = -3.68, R) >droWil1.scaffold_180701 2743212 120 - 3904529 CGCUAUUAUAACGAUCAACGGCAACUGAAAGCAAGGAGAACGCUGUCUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUA .............(((((((((..((.......))......))))).))))((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. ( -25.60, z-score = -2.39, R) >droVir3.scaffold_12875 8697087 120 - 20611582 CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGUUGCUUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUAGAUACGCUCUCUA .............((((..((((((....(....)......))))))))))((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. ( -28.70, z-score = -2.80, R) >droMoj3.scaffold_6496 11951561 120 - 26866924 CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGCUGCCUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUAGAUACACUCUCUA .............((((.((((.......(....)......))))..))))((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. ( -26.82, z-score = -2.68, R) >droGri2.scaffold_15245 11791158 120 + 18325388 CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGCUGCUUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAAUUAGAUACACUCUCUA .............(((((((((.......(....)......)))).)))))(((((((((((..((((((....))))))...(((((((........)))))))))))..))))))).. ( -29.22, z-score = -3.36, R) >consensus CGCUAUUAUAACGAUCAACGGCAACUGAACGCAAGGAGAACGCUGCUCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUA .............(((..((((.......(....)......))))...)))((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).. (-23.84 = -23.85 + 0.02)
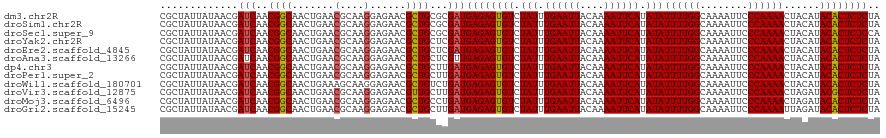
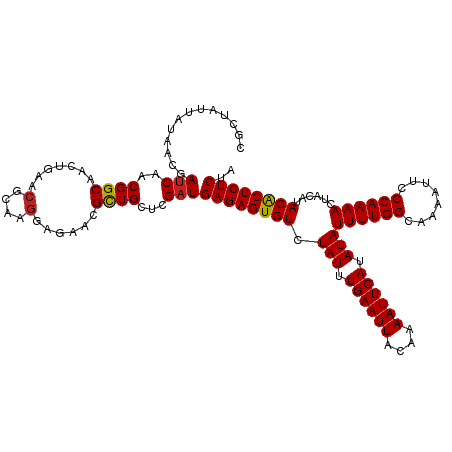
| Location | 19,823,177 – 19,823,297 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.71 |
| Shannon entropy | 0.05077 |
| G+C content | 0.37292 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -28.45 |
| Energy contribution | -28.45 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19823177 120 - 21146708 UAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGCGCAGCGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))..((((((((................)))))((.....))........))) ( -30.29, z-score = -1.79, R) >droSim1.chr2R 18375569 120 - 19596830 UAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGCGCAGCGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))..((((((((................)))))((.....))........))) ( -30.29, z-score = -1.79, R) >droSec1.super_9 3107153 120 - 3197100 UGGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGCGCAGCGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))..((((((((................)))))((.....))........))) ( -30.09, z-score = -1.38, R) >droYak2.chr2R 19800778 120 - 21139217 UAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGAGCAGCGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))..(((.(((((........(((......)))))))).)))........... ( -30.80, z-score = -2.19, R) >droEre2.scaffold_4845 21195937 120 - 22589142 UAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGAGCAGCGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))..(((.(((((........(((......)))))))).)))........... ( -30.80, z-score = -2.19, R) >droAna3.scaffold_13266 5688203 120 - 19884421 UAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAACGAGCAGCGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))(((((.(((((........(((......)))))))).)))))......... ( -33.70, z-score = -3.09, R) >dp4.chr3 1764625 120 - 19779522 UAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAGCGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))((((((((((................)))))..)))))(((((...))))) ( -30.59, z-score = -2.45, R) >droPer1.super_2 1952213 120 - 9036312 UAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAGCGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))((((((((((................)))))..)))))(((((...))))) ( -30.59, z-score = -2.45, R) >droWil1.scaffold_180701 2743212 120 + 3904529 UAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAGACAGCGUUCUCCUUGCUUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))....((.((((.......)))).))....((..(((.......)))..)). ( -29.10, z-score = -2.00, R) >droVir3.scaffold_12875 8697087 120 + 20611582 UAGAGAGCGUAUCUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAACGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..(((((.(..(((((((((((........)))))))...((((((....))))))..))))).)))))((((((((((................)))))..)))))(((((...))))) ( -26.39, z-score = -1.38, R) >droMoj3.scaffold_6496 11951561 120 + 26866924 UAGAGAGUGUAUCUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAGGCAGCGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..(((((((..(((((((((((........)))))))...((((((....))))))..)))))))))))((((((((((................))))))..))))(((((...))))) ( -32.69, z-score = -2.96, R) >droGri2.scaffold_15245 11791158 120 - 18325388 UAGAGAGUGUAUCUAAUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAGCGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..(((((((..(((((((((((........)))))))...((((((....))))))..)))))))))))((((((((((................)))))..)))))(((((...))))) ( -29.99, z-score = -2.57, R) >consensus UAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAGCGUUCUCCUUGCGUUCAGUUGCCGUUGAUCGUUAUAAUAGCG ..((((((((.....(((((((........)))))))...((((((....)))))).....))))))))((((((((((................)))))..)))))(((((...))))) (-28.45 = -28.45 + 0.00)

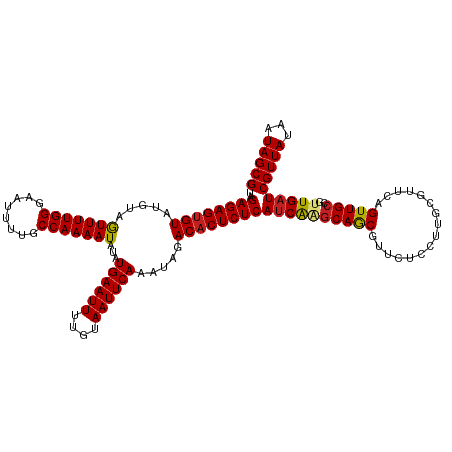
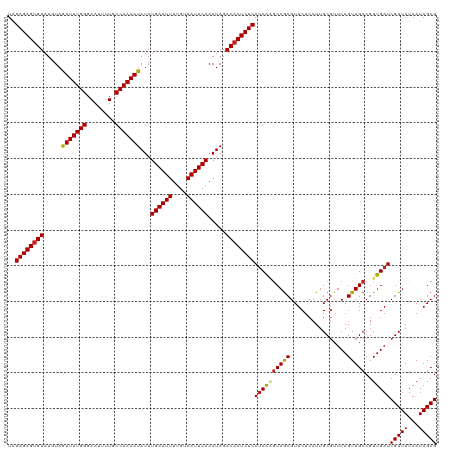
| Location | 19,823,217 – 19,823,336 |
|---|---|
| Length | 119 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.68 |
| Shannon entropy | 0.07329 |
| G+C content | 0.35690 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -24.71 |
| Energy contribution | -24.57 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19823217 119 + 21146708 CGCUGCGCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAAACUUU- .(((((.....((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........))))))......))).)).........- ( -25.60, z-score = -2.52, R) >droSim1.chr2R 18375609 119 + 19596830 CGCUGCGCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAAACUUU- .(((((.....((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........))))))......))).)).........- ( -25.60, z-score = -2.52, R) >droSec1.super_9 3107193 119 + 3197100 CGCUGCGCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCCAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAAACUUU- ...((.(((..((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).......((..(((......)))...)).))).))......- ( -24.50, z-score = -2.01, R) >droYak2.chr2R 19800818 119 + 21139217 CGCUGCUCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAAACUUU- .((((((.((.((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........))))))..)).)))).)).........- ( -25.70, z-score = -2.96, R) >droEre2.scaffold_4845 21195977 119 + 22589142 CGCUGCUCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAAACUUU- .((((((.((.((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........))))))..)).)))).)).........- ( -25.70, z-score = -2.96, R) >droAna3.scaffold_13266 5688243 119 + 19884421 CGCUGCUCGUUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAAACUUU- .((((((....((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........)))))).....)))).)).........- ( -25.60, z-score = -2.78, R) >dp4.chr3 1764665 119 + 19779522 CGCUGCUUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAAACUUU- .((((((.((.((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........))))))..)).)))).)).........- ( -25.70, z-score = -2.89, R) >droPer1.super_2 1952253 119 + 9036312 CGCUGCUUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAAACUUU- .((((((.((.((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........))))))..)).)))).)).........- ( -25.70, z-score = -2.89, R) >droWil1.scaffold_180701 2743252 120 - 3904529 CGCUGUCUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUAGCCCGGGAAAAUACUUUUAGCACGCACAAACUUUU .((((.((((.((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........))))))...)))))).)).......... ( -24.50, z-score = -2.82, R) >droVir3.scaffold_12875 8697127 119 - 20611582 CGUUGCUUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUAGAUACGCUCUCUAUUUUGGCCUGGGAAAAUACUUUUAGCACGCAUAAGCUUU- ....(((((..((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).......((.((.((((....))))..)).)).)))))...- ( -25.50, z-score = -1.47, R) >droMoj3.scaffold_6496 11951601 119 - 26866924 CGCUGCCUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUAGAUACACUCUCUAUUUUGGCCUGGGAAAAUACUUUUAGCACGCAUCAACUUU- .((.(((.(((((((((((.(((.((((((....)))))).)))((((((........))))))......)))))))).)))..)))((((((......))))))...)).........- ( -27.30, z-score = -2.77, R) >droGri2.scaffold_15245 11791198 119 + 18325388 CGCUGCUUGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAAUUAGAUACACUCUCUAUUUUGGCCUGGGAAAAUACUUUUAGCACGCACAAACUUU- .((((((.((.(((((((((((..((((((....))))))...(((((((........)))))))))))..)))))))((((((........))))))..)).)))).)).........- ( -26.30, z-score = -2.51, R) >consensus CGCUGCUCGAUGAGAGUGUCUAUUUGAAUUACAAAAUUCAUAUAUUUUGGCAAAAUUCCCAAAACUACAUACACUCUCUAUUUUGGCCCGGGAAAAUACUUUUAGCACGCACAAACUUU_ .(((((.....((((((((.(((.((((((....)))))).)))((((((........))))))......))))))))((((((........))))))......))).)).......... (-24.71 = -24.57 + -0.14)

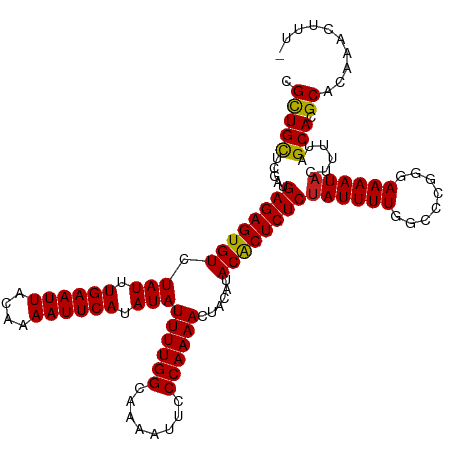
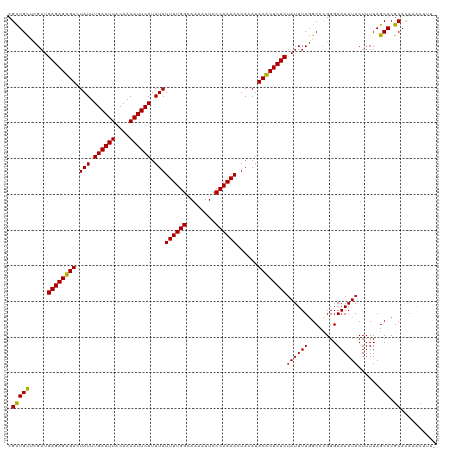
| Location | 19,823,217 – 19,823,336 |
|---|---|
| Length | 119 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.68 |
| Shannon entropy | 0.07329 |
| G+C content | 0.35690 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -27.05 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.957906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 19823217 119 - 21146708 -AAAGUUUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGCGCAGCG -...(((.(((((.(((....(......)...))).......((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))..)))))))). ( -32.20, z-score = -2.74, R) >droSim1.chr2R 18375609 119 - 19596830 -AAAGUUUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGCGCAGCG -...(((.(((((.(((....(......)...))).......((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))..)))))))). ( -32.20, z-score = -2.74, R) >droSec1.super_9 3107193 119 - 3197100 -AAAGUUUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUGGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGCGCAGCG -...(((.(((((.(((....(......)...))).......((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))..)))))))). ( -32.00, z-score = -2.27, R) >droYak2.chr2R 19800818 119 - 21139217 -AAAGUUUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGAGCAGCG -.........((.((((.....((((((........))))))((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))....)))))). ( -30.10, z-score = -2.33, R) >droEre2.scaffold_4845 21195977 119 - 22589142 -AAAGUUUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCGAGCAGCG -.........((.((((.....((((((........))))))((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))....)))))). ( -30.10, z-score = -2.33, R) >droAna3.scaffold_13266 5688243 119 - 19884421 -AAAGUUUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAACGAGCAGCG -...((((((((.((..(((.....)))..)).)).......((((((((.(((.(((((((........)))))))...((((((....)))))).))).)))))))).)))))).... ( -30.30, z-score = -2.37, R) >dp4.chr3 1764665 119 - 19779522 -AAAGUUUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAGCG -.........((.((((.....((((((........))))))((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))....)))))). ( -30.00, z-score = -2.49, R) >droPer1.super_2 1952253 119 - 9036312 -AAAGUUUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAGCG -.........((.((((.....((((((........))))))((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))....)))))). ( -30.00, z-score = -2.49, R) >droWil1.scaffold_180701 2743252 120 + 3904529 AAAAGUUUGUGCGUGCUAAAAGUAUUUUCCCGGGCUAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAGACAGCG ....(((((.((.((..(((.....)))..)).)).......((((((((.(((.(((((((........)))))))...((((((....)))))).))).))))))))..))))).... ( -29.10, z-score = -2.23, R) >droVir3.scaffold_12875 8697127 119 + 20611582 -AAAGCUUAUGCGUGCUAAAAGUAUUUUCCCAGGCCAAAAUAGAGAGCGUAUCUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAACG -...((((..((.((..(((.....)))..)).)).......(((((.(..(((((((((((........)))))))...((((((....))))))..))))).)))))...)))).... ( -25.20, z-score = -1.59, R) >droMoj3.scaffold_6496 11951601 119 + 26866924 -AAAGUUGAUGCGUGCUAAAAGUAUUUUCCCAGGCCAAAAUAGAGAGUGUAUCUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAGGCAGCG -.........((.((((.....((((((........))))))(((((((..(((((((((((........)))))))...((((((....))))))..)))))))))))....)))))). ( -29.70, z-score = -2.35, R) >droGri2.scaffold_15245 11791198 119 - 18325388 -AAAGUUUGUGCGUGCUAAAAGUAUUUUCCCAGGCCAAAAUAGAGAGUGUAUCUAAUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAGCG -.........((.((((.....((((((........))))))(((((((..(((((((((((........)))))))...((((((....))))))..)))))))))))....)))))). ( -29.40, z-score = -2.84, R) >consensus _AAAGUUUGUGCGUGCUAAAAGUAUUUUCCCGGGCCAAAAUAGAGAGUGUAUGUAGUUUUGGGAAUUUUGCCAAAAUAUAUGAAUUUUGUAAUUCAAAUAGACACUCUCAUCAAGCAGCG ..........((.((((.....((((((........))))))((((((((.....(((((((........)))))))...((((((....)))))).....))))))))....)))))). (-27.05 = -27.30 + 0.25)

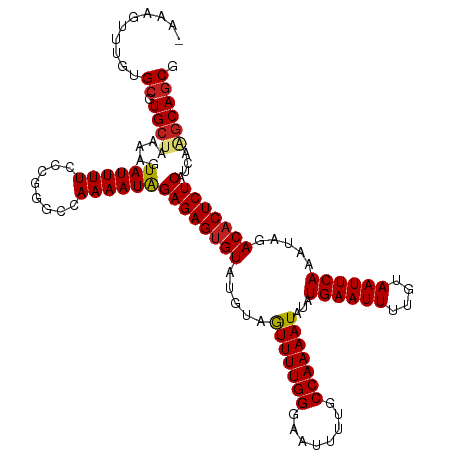
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:59 2011