| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,810,138 – 19,810,232 |
| Length | 94 |
| Max. P | 0.981931 |

| Location | 19,810,138 – 19,810,232 |
|---|---|
| Length | 94 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.50 |
| Shannon entropy | 0.60099 |
| G+C content | 0.45807 |
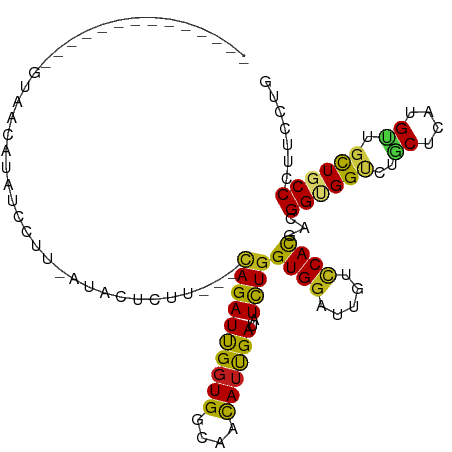
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -18.44 |
| Energy contribution | -17.43 |
| Covariance contribution | -1.01 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.981931 |
| Prediction | RNA |
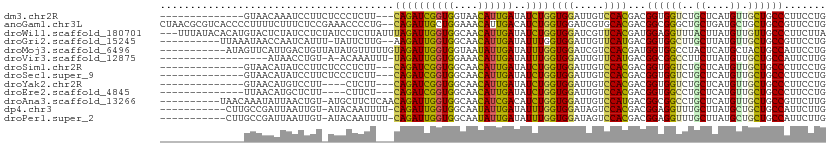
Download alignment: ClustalW | MAF
>dm3.chr2R 19810138 94 + 21146708 --------------GUAACAAAUCCUUCUCCCUCUU---CAGAUCGGUGGUAACAUUGAUAUCUGGUGGAUUGUCCACGACGGUGGUCUGCUCAUGUUGCUGCCCUUCCUG --------------......................---..((..((..(((((((.((......((((.....))))(((....)))...)))))))))..))..))... ( -24.00, z-score = -0.95, R) >anoGam1.chr3L 16661793 109 - 41284009 CUAACGCGUCACCCCUUUUCUUUCUCCGAAACCCCUG--CAGAUUGCUGGAAACAUUGACAUCUGGUGGAUCGUGCACGACGGCGGGCUGCUGAUGCUGCUGCCGUUCCUG ....(((((((((...((((.......))))..((.(--(.....)).))..............)))))..))))...((((((((.(.((....)).))))))))).... ( -28.00, z-score = 0.26, R) >droWil1.scaffold_180701 2724646 108 - 3904529 ---UUUAUACACAUGUACUCUAUCCUCUAUCCUCUUAUUUAGAUUGGUGGCAACAUUGAUAUCUGGUGGAUCGUUCACGAUGGAGGUUUACUUAUGUUGUUGCCCUUCUUA ---.....(((((((........((((((((........(((((..(((....)))..)..))))(((((...)))))))))))))......)))).)))........... ( -24.94, z-score = -1.32, R) >droGri2.scaffold_15245 6562562 98 - 18325388 ----------UUAAAUAACCAAUCAUUU-UAUUCUUG--AAGAUUGGUGGCAACAUUGAUAUUUGGUGGAUUGUUCAUGACGGUGGCUUGCUUAUGUUGCUGCCGUUCCUG ----------...(((.(((((...(((-((....))--)))((..(((....)))..))..)))))..)))......(((((..((..((....)).))..))))).... ( -25.80, z-score = -1.73, R) >droMoj3.scaffold_6496 14947174 100 + 26866924 -----------AUAGUUCAUUGACUGUUAUAUGUUUUUGUAGAUUGGUGGUAAUAUUGAUAUUUGGUGGAUCGUCCACGAUGGUGGCCUACUCAUGCUACUGCCAUUCCUG -----------((((((....))))))..............((.(((..(((.(((.((......((((..((.((.....))))..))))))))).)))..))).))... ( -20.20, z-score = 0.75, R) >droVir3.scaffold_12875 11953529 90 + 20611582 ------------------AUAACCUGU-A-ACAAAUUU-UAGAUUGGUGGAAACAUUGAUAUUUGGUGGAUUGUUCAUGACGGCGGCCUUCUUAUGUUGCUGCCAUUCUUG ------------------....(.((.-(-(((((((.-...((..(((....)))..))....)))...))))))).)..((((((...(....)..))))))....... ( -20.80, z-score = -1.02, R) >droSim1.chr2R 18361968 94 + 19596830 --------------GUAACAUAUCCUUCUCCCUCUU---CAGAUCGGUGGCAACAUUGAUAUCUGGUGGAUUGUCCACGACGGUGGUCUGCUCAUGUUGCUGCCCUUCCUG --------------(((((((......(..((((..---((((((((((....))))))..))))((((.....)))))).))..).......)))))))........... ( -27.32, z-score = -1.70, R) >droSec1.super_9 3094148 94 + 3197100 --------------GUAACAUAUCCUUCUCCCUCUU---CAGAUCGGUGGCAACAUUGAUAUCUGGUGGAUUGUCCACGACGGUGGUCUGCUCAUGUUGCUGCCCUUCCUG --------------(((((((......(..((((..---((((((((((....))))))..))))((((.....)))))).))..).......)))))))........... ( -27.32, z-score = -1.70, R) >droYak2.chr2R 19787734 90 + 21139217 --------------GUAACAUGUCCUU----CUCUU---CAGAUCGGUGGCAACAUUGAUAUCUGGUGGAUUGUCCACGACGGUGGUCUGCUCAUGUUGCUGCCCUUCCUG --------------((((((((.....----.....---((((((((((....))))))..))))(..(((..((......))..)))..).))))))))........... ( -29.40, z-score = -2.26, R) >droEre2.scaffold_4845 21182960 90 + 22589142 --------------UUAACAUGCUCUU----CUUCU---CAGAUCGGUGGCAACAUUGAUAUCUGGUGGAUUGUCCACGACGGUGGCCUGCUCAUGUUGCUGCCCUUCCUG --------------.............----.....---((((((((((....))))))..))))((((.....))))...((..((..((....)).))..))....... ( -26.10, z-score = -1.34, R) >droAna3.scaffold_13266 5674287 100 + 19884421 ----------UAACAAAUAUUAACUGU-AUGCUUCUCAACAGAUUGGUGGCAACAUCGACAUCUGGUGGAUUGUCCAUGACGGCGGCCUGCUCAUGUUGCUGCCGUUCUUG ----------.................-...........((((((((((....))))))..))))((((.....))))(((((((((..((....)).))))))))).... ( -31.60, z-score = -2.36, R) >dp4.chr3 1750902 98 + 19779522 -----------CUUGCCGAUUAAUUGU-AUACAAUUUU-CAGAUUGGUGGCAAUAUUGAUAUUUGGUGGAUAGUCCACGACGGAGGUUUGCUUAUGCUGCUGCCAUUCUUG -----------......((..(((((.-...))))).)-)(((.(((..(((.(((........((..(((..(((.....))).)))..)).))).)))..))).))).. ( -24.50, z-score = -0.93, R) >droPer1.super_2 1938475 98 + 9036312 -----------CUUGCCGAUUAAUUGU-AUACAAUUUU-CAGAUUGGUGGCAAUAUUGAUAUUUGGUGGAUAGUCCACGACGGAGGUUUGCUUAUGCUGCUGCCAUUCUUG -----------......((..(((((.-...))))).)-)(((.(((..(((.(((........((..(((..(((.....))).)))..)).))).)))..))).))).. ( -24.50, z-score = -0.93, R) >consensus ______________GUAACAUAUCCUU_AUACUCUU___CAGAUUGGUGGCAACAUUGAUAUCUGGUGGAUUGUCCACGACGGUGGUCUGCUCAUGUUGCUGCCCUUCCUG .......................................((((((((((....))))))..))))((((.....))))...((((((..((....)).))))))....... (-18.44 = -17.43 + -1.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:53 2011