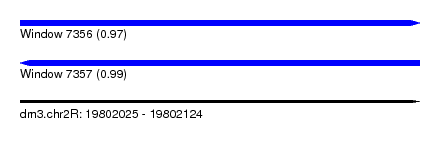
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 19,802,025 – 19,802,124 |
| Length | 99 |
| Max. P | 0.989587 |

| Location | 19,802,025 – 19,802,124 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 53.91 |
| Shannon entropy | 0.82798 |
| G+C content | 0.48685 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -14.62 |
| Energy contribution | -16.34 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.965031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
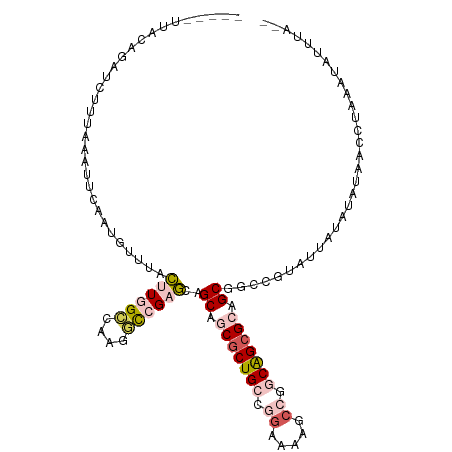
>dm3.chr2R 19802025 99 + 21146708 -----UUACAGUUUUUUA---UAUAUUUGGACUUAGGCUAAUCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGCGUUCUUAUGUAUAUCUUAUAUAUC---- -----...........((---(((((..((((....((......))((.(((.((((((.((.....)).)))))).))).))))))..)))))))...........---- ( -32.20, z-score = -1.78, R) >droSim1.chr3h_random 100618 93 - 1452968 ----------------UAAAUACAAUGGUUUCUUAGCCUAAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUUUGCGUUUCCUACAUAUAUA-- ----------------..........((((....)))).......((((((.(((((((.(((...))).))))))).))(((((......))))))))).........-- ( -34.50, z-score = -2.81, R) >dp4.Unknown_group_24 89642 111 - 109830 ACUGCUUUCAUAUCUUUUAAUUUAAAGUUUACUUGGCUAAGGCCGAGCAGCAGCGCUGCCAGAAAAGCCGUCAGCGCAGCGGCCGCAUUAUAUAUAAGCUUAAUGUUUAGU .(((((....................(((..((((((....)))))).))).((((((.(.(.....).).)))))))))))..((((((..........))))))..... ( -27.20, z-score = 0.70, R) >droPer1.super_51 391942 111 - 524598 ACUGCUUUCAUAUCUUUUAAUUUAAAGUUUACUUGGCUAAGGCCGAGCAGCAGCGCUGCCAGAAAAGCCGUCAGCGCAGCGGCCGCAUUAUAUAUAAGCUUAAUGUUUAGU .(((((....................(((..((((((....)))))).))).((((((.(.(.....).).)))))))))))..((((((..........))))))..... ( -27.20, z-score = 0.70, R) >droVir3.scaffold_13036 610849 84 + 1135500 -----UCAGAGUGUCUUACCCUCUAUCUCCAUAUGGCCAAGCCCGCACAGCCGCACUGCCGAACGCCCCCGCGGCGGAGCGGC-UUCAUA--------------------- -----...(((((((((..((.............))..)))...))))((((((.((((((..........)))))).)))))-)))...--------------------- ( -26.32, z-score = -0.88, R) >consensus _____UUACAGAUCUUUAAAUUCAAUGUUUACUUGGCCAAGGCCGAGCAGCAGCGCUGCCGGAAAAGCCGGCAGCGCAGCGGCCGUAUUAUAUAUAACCUAAAUAUUUA__ ...............................((((((....))))))..((.(((((((.((.....)).))))))).))............................... (-14.62 = -16.34 + 1.72)

| Location | 19,802,025 – 19,802,124 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 53.91 |
| Shannon entropy | 0.82798 |
| G+C content | 0.48685 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -16.44 |
| Energy contribution | -16.96 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
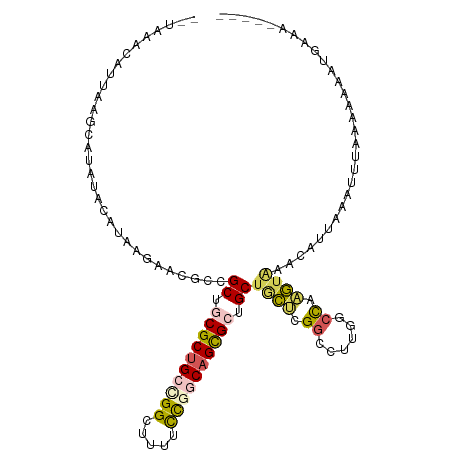
>dm3.chr2R 19802025 99 - 21146708 ----GAUAUAUAAGAUAUACAUAAGAACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUUAGCCUAAGUCCAAAUAUA---UAAAAAACUGUAA----- ----..((((((................((.(((.(((((((((.....))))))))).)))...))(((((......)))))...))))---))...........----- ( -29.90, z-score = -2.48, R) >droSim1.chr3h_random 100618 93 + 1452968 --UAUAUAUGUAGGAAACGCAAAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUAGGCUAAGAAACCAUUGUAUUUA---------------- --.(((((((.((((.((((......)))).((.(((((((.(((...))).))))))).)))))))).....((........))..)))))...---------------- ( -31.10, z-score = -1.33, R) >dp4.Unknown_group_24 89642 111 + 109830 ACUAAACAUUAAGCUUAUAUAUAAUGCGGCCGCUGCGCUGACGGCUUUUCUGGCAGCGCUGCUGCUCGGCCUUAGCCAAGUAAACUUUAAAUUAAAAGAUAUGAAAGCAGU ............((((.(((((..((((((.((.((((((.(((.....))).)))))).)).))).(((....)))..)))..((((......))))))))).))))... ( -32.60, z-score = -0.97, R) >droPer1.super_51 391942 111 + 524598 ACUAAACAUUAAGCUUAUAUAUAAUGCGGCCGCUGCGCUGACGGCUUUUCUGGCAGCGCUGCUGCUCGGCCUUAGCCAAGUAAACUUUAAAUUAAAAGAUAUGAAAGCAGU ............((((.(((((..((((((.((.((((((.(((.....))).)))))).)).))).(((....)))..)))..((((......))))))))).))))... ( -32.60, z-score = -0.97, R) >droVir3.scaffold_13036 610849 84 - 1135500 ---------------------UAUGAA-GCCGCUCCGCCGCGGGGGCGUUCGGCAGUGCGGCUGUGCGGGCUUGGCCAUAUGGAGAUAGAGGGUAAGACACUCUGA----- ---------------------..(.((-(((((.(.((((((..(.(....).)..)))))).).)).))))).)((....))...((((((......).))))).----- ( -29.90, z-score = 0.70, R) >consensus __UAAACAUUAAGCAUAUACAUAAGAACGCCGCUGCGCUGCCGGCUUUUCCGGCAGCGCUGCUGCUCGGCCUUGGCCAAGUAAACAUUAAAUUUAAAAAAAUGAAA_____ ...............................((.((((((((((.....)))))))))).))((((.((......)).))))............................. (-16.44 = -16.96 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:46:51 2011